2D protrusion but not motility predicts growth factor-induced cancer cell migration in 3D collagen
- PMID: 22665521
- PMCID: PMC3373410
- DOI: 10.1083/jcb.201201003
2D protrusion but not motility predicts growth factor-induced cancer cell migration in 3D collagen
Abstract
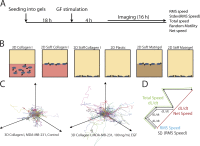
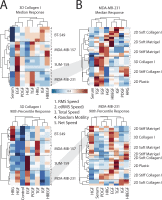
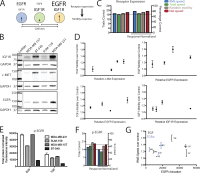
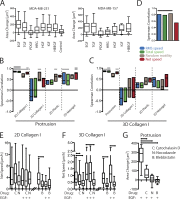
Growth factor-induced migration is a critical step in the dissemination and metastasis of solid tumors. Although differences in properties characterizing cell migration on two-dimensional (2D) substrata versus within three-dimensional (3D) matrices have been noted for particular growth factor stimuli, the 2D approach remains in more common use as an efficient surrogate, especially for high-throughput experiments. We therefore were motivated to investigate which migration properties measured in various 2D assays might be reflective of 3D migratory behavioral responses. We used human triple-negative breast cancer lines stimulated by a panel of receptor tyrosine kinase ligands relevant to mammary carcinoma progression. Whereas 2D migration properties did not correlate well with 3D behavior across multiple growth factors, we found that increased membrane protrusion elicited by growth factor stimulation did relate robustly to enhanced 3D migration properties of the MDA-MB-231 and MDA-MB-157 lines. Interestingly, we observed this to be a more reliable relationship than cognate receptor expression or activation levels across these and two additional mammary tumor lines.
Figures

References
-
- Bear J.E., Svitkina T.M., Krause M., Schafer D.A., Loureiro J.J., Strasser G.A., Maly I.V., Chaga O.Y., Cooper J.A., Borisy G.G., Gertler F.B. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109:509–521 10.1016/S0092-8674(02)00731-6 - DOI - PubMed
-
- Charafe-Jauffret E., Ginestier C., Monville F., Finetti P., Adélaïde J., Cervera N., Fekairi S., Xerri L., Jacquemier J., Birnbaum D., Bertucci F. 2006. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 25:2273–2284 10.1038/sj.onc.1209254 - DOI - PubMed
-
- Cheng N., Bhowmick N.A., Chytil A., Gorksa A.E., Brown K.A., Muraoka R., Arteaga C.L., Neilson E.G., Hayward S.W., Moses H.L. 2005. Loss of TGF-β type II receptor in fibroblasts promotes mammary carcinoma growth and invasion through upregulation of TGF-α-, MSP- and HGF-mediated signaling networks. Oncogene. 24:5053–5068 10.1038/sj.onc.1208685 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

