Noninvasive prenatal diagnosis of fetal trisomy 21 by allelic ratio analysis using targeted massively parallel sequencing of maternal plasma DNA
- PMID: 22666469
- PMCID: PMC3362548
- DOI: 10.1371/journal.pone.0038154
Noninvasive prenatal diagnosis of fetal trisomy 21 by allelic ratio analysis using targeted massively parallel sequencing of maternal plasma DNA
Abstract
Background: Plasma DNA obtained from a pregnant woman contains a mixture of maternal and fetal DNA. The fetal DNA proportion in maternal plasma is relatively consistent as determined using polymorphic genetic markers across different chromosomes in euploid pregnancies. For aneuploid pregnancies, the observed fetal DNA proportion measured using polymorphic genetic markers for the aneuploid chromosome would be perturbed. In this study, we investigated the feasibility of analyzing single nucleotide polymorphisms using targeted massively parallel sequencing to detect such perturbations in mothers carrying trisomy 21 fetuses.
Methodology/principal findings: DNA was extracted from plasma samples collected from fourteen pregnant women carrying singleton fetuses. Hybridization-based targeted sequencing was used to enrich 2 906 single nucleotide polymorphism loci on chr7, chr13, chr18 and chr21. Plasma DNA libraries with and without target enrichment were analyzed by massively parallel sequencing. Genomic DNA samples of both the mother and fetus for each case were genotyped by single nucleotide polymorphism microarray analysis. For the targeted regions, the mean sequencing depth of the enriched samples was 225-fold higher than that of the non-enriched samples. From the targeted sequencing data, the ratio between fetus-specific and shared alleles increased by approximately 2-fold on chr21 in the paternally-derived trisomy 21 case. In comparison, the ratio is decreased by approximately 11% on chr21 in the maternally-derived trisomy 21 cases but with much overlap with the ratio of the euploid cases. Computer simulation revealed the relationship between the fetal DNA proportion, the number of informative alleles and the depth of sequencing.
Conclusions/significance: Targeted massively parallel sequencing of single nucleotide polymorphism loci in maternal plasma DNA is a potential approach for trisomy 21 detection. However, the method appears to be less robust than approaches using non-polymorphism-based counting of sequence tags in plasma.
Conflict of interest statement
Figures

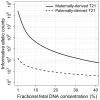
 would be 1 if the mother is carrying a euploid fetus, would become 2 if the mother is carrying a paternally-derived T21 fetus, and would become (1-f/2) if the mother is carrying a maternally-derived T21 fetus.
would be 1 if the mother is carrying a euploid fetus, would become 2 if the mother is carrying a paternally-derived T21 fetus, and would become (1-f/2) if the mother is carrying a maternally-derived T21 fetus.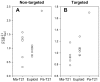
 values were calculated to differentiate the paternally- and maternally-derived T21 from the euploid fetuses in non-targeted (A) and targeted (B) sequencing data. (Ma-T21: maternally-derived T21. Pa-T21: paternally-derived T21.).
values were calculated to differentiate the paternally- and maternally-derived T21 from the euploid fetuses in non-targeted (A) and targeted (B) sequencing data. (Ma-T21: maternally-derived T21. Pa-T21: paternally-derived T21.).
References
-
- Driscoll DA, Gross S. Prenatal screening for aneuploidy. N Engl J Med. 2009;360:2556–2562. - PubMed
-
- Mujezinovic F, Alfirevic Z. Procedure-related complications of amniocentesis and chorionic villous sampling: a systematic review. Obstet Gynecol. 2007;110:687–694. - PubMed
-
- Lo YMD, Corbetta N, Chamberlain PF, Rai V, Sargent IL, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. - PubMed
-
- Tong YK, Ding CM, Chiu RWK, Gerovassili A, Chim SSC, et al. Noninvasive prenatal detection of fetal trisomy 18 by epigenetic allelic ratio analysis in maternal plasma: theoretical and empirical considerations. Clin Chem. 2006;52:2194–2202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

