Fully functionalized small-molecule probes for integrated phenotypic screening and target identification
- PMID: 22667687
- PMCID: PMC3426452
- DOI: 10.1021/ja304213w
Fully functionalized small-molecule probes for integrated phenotypic screening and target identification
Abstract
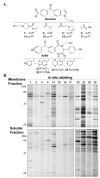
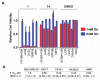
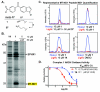
Phenotypic screening offers a powerful approach to identify small molecules that perturb complex biological processes in cells and organisms. The tendency of small molecules, however, to interact with multiple protein targets, often with moderate to weak affinities, along with the lack of straightforward technologies to characterize these interactions in living systems, has hindered efforts to understand the mechanistic basis for pharmacological activity. Here we address this challenge by creating a fully functionalized small-molecule library whose membership is endowed with: (1) one or more diversity elements to promote interactions with different protein targets in cells, (2) a photoreactive group for UV light-induced covalent cross-linking to interacting proteins, and (3) an alkyne handle for reporter tag conjugation to visualize and identify cross-linked proteins. A library member was found to inhibit cancer cell proliferation selectively under nutrient-limiting (low glucose) conditions. Quantitative chemoproteomics identified MT-ND1, an integral membrane subunit of the ∼1 MDa NADH:ubiquinone oxidoreductase (complex 1) involved in oxidative phosphorylation, as a specific target of the active probe. We further demonstrated that the active probe inhibits complex 1 activity in vitro (IC(50) = 720 nM), an effect that is known to induce cell death in low-glucose conditions. Based on this proof of principle study, we anticipate that the generation and integration of fully functionalized compound libraries into phenotypic screening programs should facilitate the discovery of bioactive probes that are amenable to accelerated target identification and mechanistic characterization using advanced chemoproteomic technologies.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

