Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels
- PMID: 22668517
- PMCID: PMC3605737
- DOI: 10.2144/000113864
Improved methodology for the affinity isolation of human protein complexes expressed at near endogenous levels
Abstract
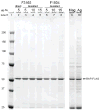
An efficient and reliable procedure for the capture of affinity-tagged proteins and associated complexes from human cell lines is reported. Through multiple optimizations, high yield and low background affinity-purifications are achieved from modest quantities of human cells expressing endogenous-level tagged proteins. Isolations of triple-FLAG and GFP-tagged fusion proteins involved in RNA metabolism are presented.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
-
- Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. - PubMed
-
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. - PubMed
-
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. - PubMed
-
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
