Low-resolution solution structures of Munc18:Syntaxin protein complexes indicate an open binding mode driven by the Syntaxin N-peptide
- PMID: 22670057
- PMCID: PMC3382502
- DOI: 10.1073/pnas.1116975109
Low-resolution solution structures of Munc18:Syntaxin protein complexes indicate an open binding mode driven by the Syntaxin N-peptide
Abstract
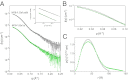
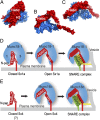
When nerve cells communicate, vesicles from one neuron fuse with the presynaptic membrane releasing chemicals that signal to the next. Similarly, when insulin binds its receptor on adipocytes or muscle, glucose transporter-4 vesicles fuse with the cell membrane, allowing glucose to be imported. These essential processes require the interaction of SNARE proteins on vesicle and cell membranes, as well as the enigmatic protein Munc18 that binds the SNARE protein Syntaxin. Here, we show that in solution the neuronal protein Syntaxin1a interacts with Munc18-1 whether or not the Syntaxin1a N-peptide is present. Conversely, the adipocyte protein Syntaxin4 does not bind its partner Munc18c unless the N-peptide is present. Solution-scattering data for the Munc18-1:Syntaxin1a complex in the absence of the N-peptide indicates that this complex adopts the inhibitory closed binding mode, exemplified by a crystal structure of the complex. However, when the N-peptide is present, the solution-scattering data indicate both Syntaxin1a and Syntaxin4 adopt extended conformations in complexes with their respective Munc18 partners. The low-resolution solution structure of the open Munc18:Syntaxin binding mode was modeled using data from cross-linking/mass spectrometry, small-angle X-ray scattering, and small-angle neutron scattering with contrast variation, indicating significant differences in Munc18:Syntaxin interactions compared with the closed binding mode. Overall, our results indicate that the neuronal Munc18-1:Syntaxin1a proteins can adopt two alternate and functionally distinct binding modes, closed and open, depending on the presence of the N-peptide, whereas Munc18c:Syntaxin4 adopts only the open binding mode.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Evidence for a conserved inhibitory binding mode between the membrane fusion assembly factors Munc18 and syntaxin in animals.J Biol Chem. 2017 Dec 15;292(50):20449-20460. doi: 10.1074/jbc.M117.811182. Epub 2017 Oct 18. J Biol Chem. 2017. PMID: 29046354 Free PMC article.
-
Revisiting interaction specificity reveals neuronal and adipocyte Munc18 membrane fusion regulatory proteins differ in their binding interactions with partner SNARE Syntaxins.PLoS One. 2017 Oct 31;12(10):e0187302. doi: 10.1371/journal.pone.0187302. eCollection 2017. PLoS One. 2017. PMID: 29088285 Free PMC article.
-
Dual modes of Munc18-1/SNARE interactions are coupled by functionally critical binding to syntaxin-1 N terminus.J Neurosci. 2007 Nov 7;27(45):12147-55. doi: 10.1523/JNEUROSCI.3655-07.2007. J Neurosci. 2007. PMID: 17989281 Free PMC article.
-
Munc18-1 as a key regulator of neurosecretion.J Neurochem. 2010 Oct;115(1):1-10. doi: 10.1111/j.1471-4159.2010.06900.x. J Neurochem. 2010. PMID: 20681955 Review.
-
The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices--guilty as charged?Annu Rev Cell Dev Biol. 2012;28:279-308. doi: 10.1146/annurev-cellbio-101011-155818. Annu Rev Cell Dev Biol. 2012. PMID: 23057743 Review.
Cited by
-
To protect or reject.Elife. 2014 Jun 17;3:e03374. doi: 10.7554/eLife.03374. Elife. 2014. PMID: 24940001 Free PMC article.
-
Evidence for a conserved inhibitory binding mode between the membrane fusion assembly factors Munc18 and syntaxin in animals.J Biol Chem. 2017 Dec 15;292(50):20449-20460. doi: 10.1074/jbc.M117.811182. Epub 2017 Oct 18. J Biol Chem. 2017. PMID: 29046354 Free PMC article.
-
Doc2b serves as a scaffolding platform for concurrent binding of multiple Munc18 isoforms in pancreatic islet β-cells.Biochem J. 2014 Dec 1;464(2):251-8. doi: 10.1042/BJ20140845. Biochem J. 2014. PMID: 25190515 Free PMC article.
-
Reconciling the regulatory role of Munc18 proteins in SNARE-complex assembly.IUCrJ. 2014 Oct 21;1(Pt 6):505-13. doi: 10.1107/S2052252514020727. eCollection 2014 Nov 1. IUCrJ. 2014. PMID: 25485130 Free PMC article. Review.
-
The nature of the Syntaxin4 C-terminus affects Munc18c-supported SNARE assembly.PLoS One. 2017 Aug 25;12(8):e0183366. doi: 10.1371/journal.pone.0183366. eCollection 2017. PLoS One. 2017. PMID: 28841669 Free PMC article.
References
-
- Jahn R, Scheller RH. SNAREs—engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. - PubMed
-
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. - PubMed
-
- Li F, et al. Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat Struct Mol Biol. 2007;14:890–896. - PubMed
-
- Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

