Suppression of premature termination codons as a therapeutic approach
- PMID: 22672057
- PMCID: PMC3432268
- DOI: 10.3109/10409238.2012.694846
Suppression of premature termination codons as a therapeutic approach
Abstract
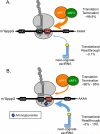
In this review, we describe our current understanding of translation termination and pharmacological agents that influence the accuracy of this process. A number of drugs have been identified that induce suppression of translation termination at in-frame premature termination codons (PTCs; also known as nonsense mutations) in mammalian cells. We discuss efforts to utilize these drugs to suppress disease-causing PTCs that result in the loss of protein expression and function. In-frame PTCs represent a genotypic subset of mutations that make up ~11% of all known mutations that cause genetic diseases, and millions of patients have diseases attributable to PTCs. Current approaches aimed at reducing the efficiency of translation termination at PTCs (referred to as PTC suppression therapy) have the goal of alleviating the phenotypic consequences of a wide range of genetic diseases. Suppression therapy is currently in clinical trials for treatment of several genetic diseases caused by PTCs, and preliminary results suggest that some patients have shown clinical improvements. While current progress is promising, we discuss various approaches that may further enhance the efficiency of this novel therapeutic approach.
Figures








References
-
- ALDENHOVEN M, BOELENS JJ, DE KONING TJ. The clinical outcome of Hurler syndrome after stem cell transplantation. Biol Blood Marrow Transplant. 2008;14:485–98. - PubMed
-
- ALI BH, AL ZA'ABI M, BLUNDEN G, NEMMAR A. Experimental gentamicin nephrotoxicity and agents that modify it: a mini-review of recent research. Basic Clin Pharmacol Toxicol. 2011;109:225–32. - PubMed
-
- ALKALAEVA EZ, PISAREV AV, FROLOVA LY, KISSELEV LL, PESTOVA TV. In Vitro Reconstitution of Eukaryotic Translation Reveals Cooperativity between Release Factors eRF1 and eRF3. Cell. 2006;125:1125–36. - PubMed
-
- AMARAL MD. Processing of CFTR: traversing the cellular maze--how much CFTR needs to go through to avoid cystic fibrosis? Pediatr Pulmonol. 2005;39:479–91. - PubMed
-
- AMRANI N, GANESAN R, KERVESTIN S, MANGUS DA, GHOSH S, JACOBSON A. A faux 3'-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
