Deciphering the genomic structure, function and evolution of carotenogenesis related phytoene synthases in grasses
- PMID: 22672222
- PMCID: PMC3413518
- DOI: 10.1186/1471-2164-13-221
Deciphering the genomic structure, function and evolution of carotenogenesis related phytoene synthases in grasses
Abstract
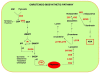
Background: Carotenoids are isoprenoid pigments, essential for photosynthesis and photoprotection in plants. The enzyme phytoene synthase (PSY) plays an essential role in mediating condensation of two geranylgeranyl diphosphate molecules, the first committed step in carotenogenesis. PSY are nuclear enzymes encoded by a small gene family consisting of three paralogous genes (PSY1-3) that have been widely characterized in rice, maize and sorghum.
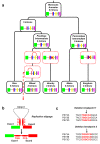
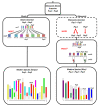
Results: In wheat, for which yellow pigment content is extremely important for flour colour, only PSY1 has been extensively studied because of its association with QTLs reported for yellow pigment whereas PSY2 has been partially characterized. Here, we report the isolation of bread wheat PSY3 genes from a Renan BAC library using Brachypodium as a model genome for the Triticeae to develop Conserved Orthologous Set markers prior to gene cloning and sequencing. Wheat PSY3 homoeologous genes were sequenced and annotated, unravelling their novel structure associated with intron-loss events and consequent exonic fusions. A wheat PSY3 promoter region was also investigated for the presence of cis-acting elements involved in the response to abscisic acid (ABA), since carotenoids also play an important role as precursors of signalling molecules devoted to plant development and biotic/abiotic stress responses. Expression of wheat PSYs in leaves and roots was investigated during ABA treatment to confirm the up-regulation of PSY3 during abiotic stress.
Conclusions: We investigated the structural and functional determinisms of PSY genes in wheat. More generally, among eudicots and monocots, the PSY gene family was found to be associated with differences in gene copy numbers, allowing us to propose an evolutionary model for the entire PSY gene family in Grasses.
Figures


Similar articles
-
PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic stress-induced root carotenogenesis.Plant Physiol. 2008 Mar;146(3):1333-45. doi: 10.1104/pp.107.111120. Epub 2007 Dec 27. Plant Physiol. 2008. PMID: 18162592 Free PMC article.
-
Functional relationships of phytoene synthase 1 alleles on chromosome 7A controlling flour colour variation in selected Australian wheat genotypes.Theor Appl Genet. 2011 Jun;123(1):95-108. doi: 10.1007/s00122-011-1569-9. Epub 2011 Mar 27. Theor Appl Genet. 2011. PMID: 21442411
-
The maize phytoene synthase gene family: overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance.Plant Physiol. 2008 Jul;147(3):1334-46. doi: 10.1104/pp.108.122119. Epub 2008 May 28. Plant Physiol. 2008. PMID: 18508954 Free PMC article.
-
Evolution of Labdane-Related Diterpene Synthases in Cereals.Plant Cell Physiol. 2020 Dec 23;61(11):1850-1859. doi: 10.1093/pcp/pcaa106. Plant Cell Physiol. 2020. PMID: 32810270 Review.
-
Evolution of root-specific carotenoid precursor pathways for apocarotenoid signal biogenesis.Plant Sci. 2015 Apr;233:1-10. doi: 10.1016/j.plantsci.2014.12.017. Epub 2014 Dec 30. Plant Sci. 2015. PMID: 25711808 Review.
Cited by
-
Differential interaction of Or proteins with the PSY enzymes in saffron.Sci Rep. 2020 Jan 17;10(1):552. doi: 10.1038/s41598-020-57480-2. Sci Rep. 2020. PMID: 31953512 Free PMC article.
-
Differential Regulation of Phytoene Synthase PSY1 During Fruit Carotenogenesis in Cultivated and Wild Tomato Species (Solanum section Lycopersicon).Plants (Basel). 2020 Sep 9;9(9):1169. doi: 10.3390/plants9091169. Plants (Basel). 2020. PMID: 32916928 Free PMC article.
-
Characterization and Expression Analysis of Phytoene Synthase from Bread Wheat (Triticum aestivum L.).PLoS One. 2016 Oct 3;11(10):e0162443. doi: 10.1371/journal.pone.0162443. eCollection 2016. PLoS One. 2016. PMID: 27695116 Free PMC article.
-
Genetic and molecular bases of yield-associated traits: a translational biology approach between rice and wheat.Theor Appl Genet. 2014 Jul;127(7):1463-89. doi: 10.1007/s00122-014-2332-9. Epub 2014 Jun 10. Theor Appl Genet. 2014. PMID: 24913362 Review.
-
Phytoene Synthase: The Key Rate-Limiting Enzyme of Carotenoid Biosynthesis in Plants.Front Plant Sci. 2022 Apr 12;13:884720. doi: 10.3389/fpls.2022.884720. eCollection 2022. Front Plant Sci. 2022. PMID: 35498681 Free PMC article. Review.
References
-
- Britton G, Liaaen-Jensen S, Pfander H. Carotenoids Handbook. Birkhäuser Verlag, Basel; 2004.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

