Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems
- PMID: 22672726
- PMCID: PMC3443267
- DOI: 10.1111/j.1462-2920.2012.02794.x
Post-transcriptional regulation on a global scale: form and function of Csr/Rsm systems
Abstract
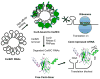
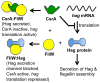
Originally described as a repressor of gene expression in the stationary phase of growth, CsrA (RsmA) regulates primary and secondary metabolic pathways, biofilm formation, motility, virulence circuitry of pathogens, quorum sensing and stress response systems by binding to conserved sequences in its target mRNAs and altering their translation and/or turnover. While the binding of CsrA to RNA is understood at an atomic level, new mechanisms of gene activation and repression by this protein are still emerging. In the γ-proteobacteria, small non-coding RNAs (sRNAs) use molecular mimicry to sequester multiple CsrA dimers away from mRNA. In contrast, the FliW protein of Bacillus subtilis inhibits CsrA activity by binding to this protein, thereby establishing a checkpoint in flagellum morphogenesis. Turnover of CsrB and CsrC sRNAs in Escherichia coli requires a specificity protein of the GGDEF-EAL domain superfamily, CsrD, in addition to the housekeeping nucleases RNase E and PNPase. The Csr system of E. coli contains extensive autoregulatory circuitry, which governs the expression and activity of CsrA. Interaction of the Csr system with transcriptional regulatory networks results in a variety of complex response patterns. This minireview will highlight basic principles and new insights into the workings of these complex eubacterial regulatory systems.
© 2012 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures


Similar articles
-
Regulation of bacterial virulence by Csr (Rsm) systems.Microbiol Mol Biol Rev. 2015 Jun;79(2):193-224. doi: 10.1128/MMBR.00052-14. Microbiol Mol Biol Rev. 2015. PMID: 25833324 Free PMC article. Review.
-
Circuitry Linking the Catabolite Repression and Csr Global Regulatory Systems of Escherichia coli.J Bacteriol. 2016 Oct 7;198(21):3000-3015. doi: 10.1128/JB.00454-16. Print 2016 Nov 1. J Bacteriol. 2016. PMID: 27551019 Free PMC article.
-
Examination of Csr regulatory circuitry using epistasis analysis with RNA-seq (Epi-seq) confirms that CsrD affects gene expression via CsrA, CsrB and CsrC.Sci Rep. 2018 Mar 29;8(1):5373. doi: 10.1038/s41598-018-23713-8. Sci Rep. 2018. PMID: 29599472 Free PMC article.
-
Antagonistic control of the turnover pathway for the global regulatory sRNA CsrB by the CsrA and CsrD proteins.Nucleic Acids Res. 2016 Sep 19;44(16):7896-910. doi: 10.1093/nar/gkw484. Epub 2016 May 27. Nucleic Acids Res. 2016. PMID: 27235416 Free PMC article.
-
Regulation of host-pathogen interactions via the post-transcriptional Csr/Rsm system.Curr Opin Microbiol. 2018 Feb;41:58-67. doi: 10.1016/j.mib.2017.11.022. Epub 2017 Dec 5. Curr Opin Microbiol. 2018. PMID: 29207313 Review.
Cited by
-
Regulation of bacterial virulence by Csr (Rsm) systems.Microbiol Mol Biol Rev. 2015 Jun;79(2):193-224. doi: 10.1128/MMBR.00052-14. Microbiol Mol Biol Rev. 2015. PMID: 25833324 Free PMC article. Review.
-
Multitier regulation of the E. coli extreme acid stress response by CsrA.J Bacteriol. 2024 Apr 18;206(4):e0035423. doi: 10.1128/jb.00354-23. Epub 2024 Feb 6. J Bacteriol. 2024. PMID: 38319100 Free PMC article.
-
Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo.EMBO J. 2016 May 2;35(9):991-1011. doi: 10.15252/embj.201593360. Epub 2016 Apr 4. EMBO J. 2016. PMID: 27044921 Free PMC article.
-
Structural basis for the CsrA-dependent modulation of translation initiation by an ancient regulatory protein.Proc Natl Acad Sci U S A. 2016 Sep 6;113(36):10168-73. doi: 10.1073/pnas.1602425113. Epub 2016 Aug 22. Proc Natl Acad Sci U S A. 2016. PMID: 27551070 Free PMC article.
-
The Csr system regulates genome-wide mRNA stability and transcription and thus gene expression in Escherichia coli.Sci Rep. 2016 Apr 26;6:25057. doi: 10.1038/srep25057. Sci Rep. 2016. PMID: 27112822 Free PMC article.
References
-
- Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, Silva IJ, Viegas SC. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 2010;34:883–923. - PubMed
-
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. - PubMed
-
- Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol. 2002;44:1599–1610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

