Large-scale analysis of microRNA evolution
- PMID: 22672736
- PMCID: PMC3497579
- DOI: 10.1186/1471-2164-13-218
Large-scale analysis of microRNA evolution
Abstract
Background: In animals, microRNAs (miRNA) are important genetic regulators. Animal miRNAs appear to have expanded in conjunction with an escalation in complexity during early bilaterian evolution. Their small size and high-degree of similarity makes them challenging for phylogenetic approaches. Furthermore, genomic locations encoding miRNAs are not clearly defined in many species. A number of studies have looked at the evolution of individual miRNA families. However, we currently lack resources for large-scale analysis of miRNA evolution.
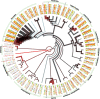
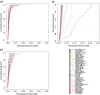
Results: We addressed some of these issues in order to analyse the evolution of miRNAs. We perform syntenic and phylogenetic analysis for miRNAs from 80 animal species. We present synteny maps, phylogenies and functional data for miRNAs across these species. These data represent the basis of our analyses and also act as a resource for the community.
Conclusions: We use these data to explore the distribution of miRNAs across phylogenetic space, characterise their birth and death, and examine functional relationships between miRNAs and other genes. These data confirm a number of previously reported findings on a larger scale and also offer novel insights into the evolution of the miRNA repertoire in animals, and it's genomic organization.
Figures


References
-
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

