Timeless preserves telomere length by promoting efficient DNA replication through human telomeres
- PMID: 22672906
- PMCID: PMC3383593
- DOI: 10.4161/cc.20810
Timeless preserves telomere length by promoting efficient DNA replication through human telomeres
Abstract
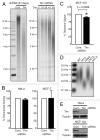
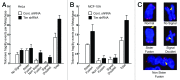
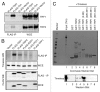
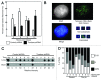
A variety of telomere protection programs are utilized to preserve telomere structure. However, the complex nature of telomere maintenance remains elusive. The Timeless protein associates with the replication fork and is thought to support efficient progression of the replication fork through natural impediments, including replication fork block sites. However, the mechanism by which Timeless regulates such genomic regions is not understood. Here, we report the role of Timeless in telomere length maintenance. We demonstrate that Timeless depletion leads to telomere shortening in human cells. This length maintenance is independent of telomerase, and Timeless depletion causes increased levels of DNA damage, leading to telomere aberrations. We also show that Timeless is associated with Shelterin components TRF1 and TRF2. Timeless depletion slows telomere replication in vitro, and Timeless-depleted cells fail to maintain TRF1-mediated accumulation of replisome components at telomeric regions. Furthermore, telomere replication undergoes a dramatic delay in Timeless-depleted cells. These results suggest that Timeless functions together with TRF1 to prevent fork collapse at telomere repeat DNA and ensure stable maintenance of telomere length and integrity.
Figures



Comment in
-
Timeless tunes: replicating happy endings.Cell Cycle. 2012 Aug 15;11(16):2977-8. doi: 10.4161/cc.21530. Epub 2012 Aug 9. Cell Cycle. 2012. PMID: 22874596 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
