Major role of positive selection in the evolution of conservative segments of Drosophila proteins
- PMID: 22673359
- PMCID: PMC3396909
- DOI: 10.1098/rspb.2012.0776
Major role of positive selection in the evolution of conservative segments of Drosophila proteins
Abstract
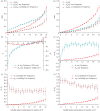
Slow evolution of conservative segments of coding and non-coding DNA is caused by the action of negative selection, which removes new mutations. However, the mode of selection that affects the few substitutions that do occur within such segments remains unclear. Here, we show that the fraction of allele replacements that were driven by positive selection, and the strength of this selection, is the highest within the conservative segments of Drosophila protein-coding genes. The McDonald-Kreitman test, applied to the data on variation in Drosophila melanogaster and in Drosophila simulans, indicates that within the most conservative protein segments, approximately 72 per cent (approx. 80%) of allele replacements were driven by positive selection, as opposed to only approximately 44 per cent (approx. 53%) at rapidly evolving segments. Data on multiple non-synonymous substitutions at a codon lead to the same conclusion and additionally indicate that positive selection driving allele replacements at conservative sites is the strongest, as it accelerates evolution by a factor of approximately 40, as opposed to a factor of approximately 5 at rapidly evolving sites. Thus, random drift plays only a minor role in the evolution of conservative DNA segments, and those relatively rare allele replacements that occur within such segments are mostly driven by substantial positive selection.
Figures


References
-
- Kimura M. 1985. The neutral theory of molecular evolution. Cambridge, UK: Cambridge University Press
-
- Yang Z., Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 19, 908–917 10.1093/oxfordjournals.molbev.a004148 (doi:10.1093/oxfordjournals.molbev.a004148) - DOI - PubMed
-
- Zhang J., Nielsen R., Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22, 2472–2479 10.1093/molbev/msi237 (doi:10.1093/molbev/msi237) - DOI - PubMed
-
- Studer R. A., Penel S., Duret L., Robinson-Rechavi M. 2008. Pervasive positive selection on duplicated and nonduplicated vertebrate protein coding genes. Genome Res. 18, 1393–1402 10.1101/gr.076992.108 (doi:10.1101/gr.076992.108) - DOI - PMC - PubMed
-
- Wang H.-Y., Tang H., Shen C.-K. J., Wu C.-I. 2003. Rapidly evolving genes in human. I. The glycophorins and their possible role in evading malaria parasites. Mol. Biol. Evol. 20, 1795–1804 10.1093/molbev/msg185 (doi:10.1093/molbev/msg185) - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
