Effects of ADARs on small RNA processing pathways in C. elegans
- PMID: 22673872
- PMCID: PMC3409262
- DOI: 10.1101/gr.134841.111
Effects of ADARs on small RNA processing pathways in C. elegans
Abstract
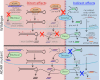
Adenosine deaminases that act on RNA (ADARs) are RNA editing enzymes that convert adenosine to inosine in double-stranded RNA (dsRNA). To evaluate effects of ADARs on small RNAs that derive from dsRNA precursors, we performed deep-sequencing, comparing small RNAs from wild-type and ADAR mutant Caenorhabditis elegans. While editing in small RNAs was rare, at least 40% of microRNAs had altered levels in at least one ADAR mutant strain, and miRNAs with significantly altered levels had mRNA targets with correspondingly affected levels. About 40% of siRNAs derived from endogenous genes (endo-siRNAs) also had altered levels in at least one mutant strain, including 63% of Dicer-dependent endo-siRNAs. The 26G class of endo-siRNAs was significantly affected by ADARs, and many altered 26G loci had intronic reads and histone modifications associated with transcriptional silencing. Our data indicate that ADARs, through both direct and indirect mechanisms, are important for maintaining wild-type levels of many small RNAs in C. elegans.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
