Efficient determination of purine metabolites in brain tissue and serum by high-performance liquid chromatography with electrochemical and UV detection
- PMID: 22674671
- PMCID: PMC9979337
- DOI: 10.1002/bmc.2760
Efficient determination of purine metabolites in brain tissue and serum by high-performance liquid chromatography with electrochemical and UV detection
Abstract
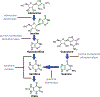
The purine metabolic pathway has been implicated in neurodegeneration and neuroprotection. High-performance liquid chromatography (HPLC) is widely used to determine purines and metabolites. However, methods for analysis of multiple purines in a single analysis have not been standardized, especially in brain tissue. We report the development and validation of a reversed-phase HPLC method combining electrochemical and UV detection after a short gradient run to measure seven purine metabolites (adenosine, guanosine, inosine, guanine, hypoxanthine, xanthine and urate) from the entire purine metabolic pathway. The limit of detection (LoD) for each analyte was determined. The LoD using UV absorption was 0.001 mg/dL for hypoxanthine (Hyp), inosine (Ino), guanosine (Guo) and adenosine (Ado), and those using coulometric electrodes were 0.001 mg/dL for guanine (Gua), 0.0001 mg/dL for urate (UA) and 0.0005 mg/dL for xanthine (Xan). The intra- and inter-day coefficient of variance was generally <8%. Using this method, we determined basal levels of these metabolites in mouse brain and serum, as well as in post-mortem human brain. Peak identities were confirmed by enzyme degradation. Spike recovery was performed to assess accuracy. All recoveries fell within 80-120%. Our HPLC method provides a sensitive, rapid, reproducible and low-cost method for determining multiple purine metabolites in a single analysis in serum and brain specimens.
Copyright © 2012 John Wiley & Sons, Ltd.
Figures



Similar articles
-
Ultra-high performance liquid chromatography with ultraviolet and tandem mass spectrometry for simultaneous determination of metabolites in purine pathway of rat plasma.J Chromatogr B Analyt Technol Biomed Life Sci. 2016 Nov 15;1036-1037:84-92. doi: 10.1016/j.jchromb.2016.09.023. Epub 2016 Sep 28. J Chromatogr B Analyt Technol Biomed Life Sci. 2016. PMID: 27736691
-
Simultaneous determination of 16 purine derivatives in urinary calculi by gradient reversed-phase high-performance liquid chromatography with UV detection.J Chromatogr B Analyt Technol Biomed Life Sci. 2005 May 25;819(2):229-35. doi: 10.1016/j.jchromb.2004.11.013. J Chromatogr B Analyt Technol Biomed Life Sci. 2005. PMID: 15833286
-
Metabolic signatures of metabolites of the purine degradation pathway in human plasma using HILIC UHPLC-HRMS.J Pharm Biomed Anal. 2024 Dec 15;251:116451. doi: 10.1016/j.jpba.2024.116451. Epub 2024 Aug 26. J Pharm Biomed Anal. 2024. PMID: 39217702
-
[The influence of some trace elements on bioaccumulation in tissues and bioenergetic metabolism of the edible snail Helix aspersa maxima as determined by HPLC of purine derivatives].Ann Acad Med Stetin. 2003;49:63-77. Ann Acad Med Stetin. 2003. PMID: 15552840 Polish.
-
Total purine and purine base content of common foodstuffs for facilitating nutritional therapy for gout and hyperuricemia.Biol Pharm Bull. 2014;37(5):709-21. doi: 10.1248/bpb.b13-00967. Epub 2014 Feb 20. Biol Pharm Bull. 2014. PMID: 24553148 Review.
Cited by
-
Protection of dopaminergic cells by urate requires its accumulation in astrocytes.J Neurochem. 2012 Oct;123(1):172-81. doi: 10.1111/j.1471-4159.2012.07820.x. Epub 2012 Aug 22. J Neurochem. 2012. PMID: 22671773 Free PMC article.
-
The renal phenotype of allopurinol-treated HPRT-deficient mouse.PLoS One. 2017 Mar 10;12(3):e0173512. doi: 10.1371/journal.pone.0173512. eCollection 2017. PLoS One. 2017. PMID: 28282408 Free PMC article.
-
Continuous monitoring of adenosine and its metabolites using microdialysis coupled to microchip electrophoresis with amperometric detection.Anal Methods. 2018 Aug 14;10(30):3737-3744. doi: 10.1039/C8AY01041B. Epub 2018 Jul 13. Anal Methods. 2018. PMID: 31579297 Free PMC article.
-
Postmortem brain levels of urate and precursors in Parkinson's disease and related disorders.Neurodegener Dis. 2013;12(4):189-98. doi: 10.1159/000346370. Epub 2013 Feb 28. Neurodegener Dis. 2013. PMID: 23467193 Free PMC article.
-
Disrupted and transgenic urate oxidase alter urate and dopaminergic neurodegeneration.Proc Natl Acad Sci U S A. 2013 Jan 2;110(1):300-5. doi: 10.1073/pnas.1217296110. Epub 2012 Dec 17. Proc Natl Acad Sci U S A. 2013. PMID: 23248282 Free PMC article.
References
-
- Ascherio A, LeWitt PA, Xu K, Eberly S, Watts A, Matson WR, Marras C, Kieburtz K, Rudolph A, Bogdanov MB, Schwid SR, Tennis M, Tanner CM, Beal MF, Lang AE, Oakes D, Fahn S, Shoulson I. Schwarzschild MA and Parkinson Study Group DATATOP Investigators. Urate as a predictor of the rate of clinical decline in Parkinson disease. Archives of Neurology 2009; 66(12): 1460–1468. - PMC - PubMed
-
- Bakay B, Nissinen E and Sweetman L. Analysis of radioactive and nonradioactive purine bases, nucleosides, and nucleotides by high-speed chromatography on a single column. Analytical Biochemistry 1978; 86(1): 65–77. - PubMed
-
- Burnstock G Purinergic signalling and disorders of the central nervous system. Nature Reviews. Drug Discovery 2008; 7: 575–590. - PubMed
-
- Gomez G and Sitkovsky MV. Differential requirement for A2a and A3 adenosine receptors for the protective effect of inosine in vivo. Blood 2003; 102(13): 4472–4478. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

