Re-engineering electrochemical biosensors to narrow or extend their useful dynamic range
- PMID: 22674785
- PMCID: PMC3482547
- DOI: 10.1002/anie.201202204
Re-engineering electrochemical biosensors to narrow or extend their useful dynamic range
Abstract
Here we demonstrate two convenient methods to extend and narrow the useful dynamic range of a model electrochemical DNA sensor. We did so by combining DNA probes of different target affinities but with similar specificity on the same electrode. We were able to achieve an extended dynamic response spanning 3 orders of magnitude in target concentration. Using a different strategy we have also narrowed the useful dynamic range of an E-DNA sensor to only an 8-fold range of target concentrations.
Figures
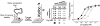


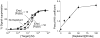

References
-
- Bullen RA, Arnot TC, Lakeman JB, Walsh FC. Biosens Bioelectron. 2006;21:2015–2045. - PubMed
-
- Privman V, Pedrosa V, Melnikov D, Pita M, Simonian A, Katz E. Biosens Bioelectron. 2009;25:695–701. - PubMed
- Wang J, Katz E. Isr J Chem. 2011;51:141–150.
-
- Koshland DE. The molecular basis for enzyme regulation. Vol. 1. Academic Press; New York: 1970.
- Goldbeter A, Koshland DE. Proc Natl Acad Sci USA. 1981;78:6840–6844. - PMC - PubMed
- Koshland DE, Goldbeter A, Stock JB. Science. 1982;217:220–225. - PubMed
- Ferrell JE. Trends Biochem Sci. 1996;21:460–466. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

