Infectious endogenous retroviruses in cats and emergence of recombinant viruses
- PMID: 22674983
- PMCID: PMC3421742
- DOI: 10.1128/JVI.00280-12
Infectious endogenous retroviruses in cats and emergence of recombinant viruses
Abstract
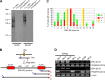
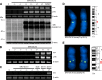
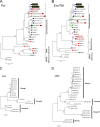
Endogenous retroviruses (ERVs) comprise a significant percentage of the mammalian genome, and it is poorly understood whether they will remain as inactive genomes or emerge as infectious retroviruses. Although several types of ERVs are present in domestic cats, infectious ERVs have not been demonstrated. Here, we report a previously uncharacterized class of endogenous gammaretroviruses, termed ERV-DCs, that is present and hereditary in the domestic cat genome. We have characterized a subset of ERV-DC proviral clones, which are numbered according to their genomic insertions. One of these, ERV-DC10, located in the q12-q21 region on chromosome C1, is an infectious gammaretrovirus capable of infecting a broad range of cells, including human. Our studies indicate that ERV-DC10 entered the genome of domestic cats in the recent past and appeared to translocate to or reintegrate at a distinct locus as infectious ERV-DC18. Insertional polymorphism analysis revealed that 92 of 244 domestic cats had ERV-DC10 on a homozygous or heterozygous locus. ERV-DC-like sequences were found in primate and rodent genomes, suggesting that these ERVs, and recombinant viruses such as RD-114 and BaEV, originated from an ancestor of ERV-DC. We also found that a novel recombinant virus, feline leukemia virus subgroup D (FeLV-D), was generated by ERV-DC env transduction into feline leukemia virus in domestic cats. Our results indicate that ERV-DCs behave as donors and/or acceptors in the generation of infectious, recombinant viruses. The presence of such infectious endogenous retroviruses, which could be harmful or beneficial to the host, may affect veterinary medicine and public health.
Figures




References
-
- Benveniste RE, Todaro GJ. 1974. Evolution of C-type viral genes: inheritance of exogenously acquired viral genes. Nature 252:456–459 - PubMed
-
- Benveniste RE, Sherr CJ, Todaro GJ. 1975. Evolution of type C viral genes: origin of feline leukemia virus. Science 190:886–888 - PubMed
-
- Beyer W, Mohring R, Drescher B, Notzel U, Rosenthal S. 1987. Molecular cloning of an endogenous cat retroviral element (ECE 1)–a recombinant between RD-114 and FeLV-related sequences. Brief report. Arch. Virol. 96:297–301 - PubMed
-
- Boeke JD, Stoye JP. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements, p 343–436 In Coffin JM, Hughes SH, Varmus HE. (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY - PubMed
-
- Bonner TI, Todaro GJ. 1979. Carnivores have sequences in their cellular DNA distantly related to the primate endogenous virus, MAC-1. Virology 94:224–227 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

