Hepatitis C virus infection alters P-body composition but is independent of P-body granules
- PMID: 22674998
- PMCID: PMC3421735
- DOI: 10.1128/JVI.07167-11
Hepatitis C virus infection alters P-body composition but is independent of P-body granules
Abstract
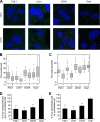
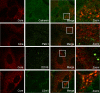
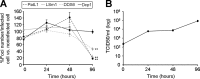
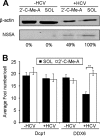
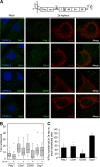
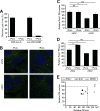
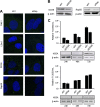
Processing bodies (P-bodies) are highly dynamic cytoplasmic granules conserved among eukaryotes. They are present under normal growth conditions and contain translationally repressed mRNAs together with proteins from the mRNA decay and microRNA (miRNA) machineries. We have previously shown that the core P-body components PatL1, LSm1, and DDX6 (Rck/p54) are required for hepatitis C virus (HCV) RNA replication; however, how HCV infection affects P-body granules and whether P-body granules per se influence the HCV life cycle remain unresolved issues. Here we show that HCV infection alters P-body composition by specifically changing the localization pattern of P-body components that are required for HCV replication. This effect was not related to an altered expression level of these components and could be reversed by inhibiting HCV replication with a polymerase inhibitor. Similar observations were obtained with a subgenomic replicon that supports only HCV translation and replication, indicating that these early steps of the HCV life cycle trigger the P-body alterations. Finally, P-body disruption by Rap55 depletion did not affect viral titers or HCV protein levels, demonstrating that the localization of PatL1, LSm1, and DDX6 in P-bodies is not required for their function on HCV. Thus, the HCV-induced changes on P-bodies are mechanistically linked to the function of specific P-body components in HCV RNA translation and replication; however, the formation of P-body granules is not required for HCV infection.
Figures

Similar articles
-
DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not for internal ribosome entry site-directed translation.J Virol. 2010 Jul;84(13):6810-24. doi: 10.1128/JVI.00397-10. Epub 2010 Apr 14. J Virol. 2010. PMID: 20392846 Free PMC article.
-
Hepatitis C virus hijacks P-body and stress granule components around lipid droplets.J Virol. 2011 Jul;85(14):6882-92. doi: 10.1128/JVI.02418-10. Epub 2011 May 4. J Virol. 2011. PMID: 21543503 Free PMC article.
-
Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates.Proc Natl Acad Sci U S A. 2009 Aug 11;106(32):13517-22. doi: 10.1073/pnas.0906413106. Epub 2009 Jul 23. Proc Natl Acad Sci U S A. 2009. PMID: 19628699 Free PMC article.
-
Hepatitis C virus plays with fire and yet avoids getting burned. A review for clinicians on processing bodies and stress granules.Liver Int. 2018 Mar;38(3):388-398. doi: 10.1111/liv.13541. Epub 2017 Sep 4. Liver Int. 2018. PMID: 28782251 Review.
-
Mammalian stress granules and processing bodies.Methods Enzymol. 2007;431:61-81. doi: 10.1016/S0076-6879(07)31005-7. Methods Enzymol. 2007. PMID: 17923231 Review.
Cited by
-
The P body protein LSm1 contributes to stimulation of hepatitis C virus translation, but not replication, by microRNA-122.Nucleic Acids Res. 2014 Jan;42(2):1257-69. doi: 10.1093/nar/gkt941. Epub 2013 Oct 18. Nucleic Acids Res. 2014. PMID: 24141094 Free PMC article.
-
P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication.Virology. 2013 Feb 5;436(1):1-7. doi: 10.1016/j.virol.2012.09.041. Epub 2012 Oct 24. Virology. 2013. PMID: 23102969 Free PMC article.
-
Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses.Virology. 2015 May;479-480:457-74. doi: 10.1016/j.virol.2015.03.001. Epub 2015 Mar 26. Virology. 2015. PMID: 25818028 Free PMC article. Review.
-
Who Regulates Whom? An Overview of RNA Granules and Viral Infections.Viruses. 2016 Jun 28;8(7):180. doi: 10.3390/v8070180. Viruses. 2016. PMID: 27367717 Free PMC article. Review.
-
Screening of small molecules affecting mammalian P-body assembly uncovers links with diverse intracellular processes and organelle physiology.RNA Biol. 2013 Nov;10(11):1661-9. doi: 10.4161/rna.26851. RNA Biol. 2013. PMID: 24418890 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

