Re-splicing of mature mRNA in cancer cells promotes activation of distant weak alternative splice sites
- PMID: 22675076
- PMCID: PMC3439910
- DOI: 10.1093/nar/gks520
Re-splicing of mature mRNA in cancer cells promotes activation of distant weak alternative splice sites
Abstract
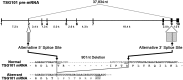
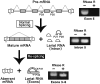
Transcripts of the human tumor susceptibility gene 101 (TSG101) are aberrantly spliced in many cancers. A major aberrant splicing event on the TSG101 pre-mRNA involves joining of distant alternative 5' and 3' splice sites within exon 2 and exon 9, respectively, resulting in the extensive elimination of the mRNA. The estimated strengths of the alternative splice sites are much lower than those of authentic splice sites. We observed that the equivalent aberrant mRNA could be generated from an intron-less TSG101 gene expressed ectopically in breast cancer cells. Remarkably, we identified a pathway-specific endogenous lariat RNA consisting solely of exonic sequences, predicted to be generated by a re-splicing between exon 2 and exon 9 on the spliced mRNA. Our results provide evidence for a two-step splicing pathway in which the initial constitutive splicing removes all 14 authentic splice sites, thereby bringing the weak alternative splice sites into close proximity. We also demonstrate that aberrant multiple-exon skipping of the fragile histidine triad (FHIT) pre-mRNA in cancer cells occurs via re-splicing of spliced FHIT mRNA. The re-splicing of mature mRNA can potentially generate mutation-independent diversity in cancer transcriptomes. Conversely, a mechanism may exist in normal cells to prevent potentially deleterious mRNA re-splicing events.
Figures





References
-
- Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. - PubMed
-
- Kalnina Z, Zayakin P, Silina K, Line A. Alterations of pre-mRNA splicing in cancer. Genes Chromosomes Cancer. 2005;42:342–357. - PubMed
-
- Pajares MJ, Ezponda T, Catena R, Calvo A, Pio R, Montuenga LM. Alternative splicing: an emerging topic in molecular and clinical oncology. Lancet Oncol. 2007;8:349–357. - PubMed
-
- Li L, Cohen SN. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell. 1996;85:319–329. - PubMed
-
- Slagsvold T, Pattni K, Malerod L, Stenmark H. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 2006;16:317–326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

