Modelling cell motility and chemotaxis with evolving surface finite elements
- PMID: 22675164
- PMCID: PMC3479903
- DOI: 10.1098/rsif.2012.0276
Modelling cell motility and chemotaxis with evolving surface finite elements
Abstract
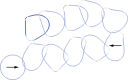
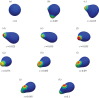
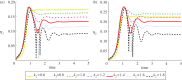
We present a mathematical and a computational framework for the modelling of cell motility. The cell membrane is represented by an evolving surface, with the movement of the cell determined by the interaction of various forces that act normal to the surface. We consider external forces such as those that may arise owing to inhomogeneities in the medium and a pressure that constrains the enclosed volume, as well as internal forces that arise from the reaction of the cells' surface to stretching and bending. We also consider a protrusive force associated with a reaction-diffusion system (RDS) posed on the cell membrane, with cell polarization modelled by this surface RDS. The computational method is based on an evolving surface finite-element method. The general method can account for the large deformations that arise in cell motility and allows the simulation of cell migration in three dimensions. We illustrate applications of the proposed modelling framework and numerical method by reporting on numerical simulations of a model for eukaryotic chemotaxis and a model for the persistent movement of keratocytes in two and three space dimensions. Movies of the simulated cells can be obtained from http://homepages.warwick.ac.uk/∼maskae/CV_Warwick/Chemotaxis.html.
Figures











References
-
- Bray D. 2001. Cell movements: from molecules to motility. London, UK: Routledge
-
- Mogilner A. 2009. Mathematics of cell motility: have we got its number? J. Math. Biol. 58, 105–13410.1007/s00285-008-0182-2 (doi:10.1007/s00285-008-0182-2) - DOI - DOI - PMC - PubMed
-
- Jilkine A., Edelstein-Keshet L. 2011. A comparison of mathematical models for polarization of single eukaryotic cells in response to guided cues. PLoS Comput. Biol. 7, e10011210.10.1371/journal.pcbi.1001121 (doi:10.1371/journal.pcbi.1001121) - DOI - DOI - PMC - PubMed
-
- Del Alamo J. C., Meili R., Alonso-Latorre B., Rodríguez-Rodríguez J., Aliseda A., Firtel R. A., Lasheras J. C. 2007. Spatio-temporal analysis of eukaryotic cell motility by improved force cytometry. Proc. Natl Acad. Sci. USA 104, 13 343 – 13 34810.1073/pnas.0705815104 (doi:10.1073/pnas.0705815104) - DOI - DOI - PMC - PubMed
-
- Lombardi M. L., Knecht D. A., Dembo M., Lee J. 2007. Traction force microscopy in Dictyostelium reveals distinct roles for myosin II motor and actin-crosslinking activity in polarized cell movement. J. Cell Sci. 120, 1624–163410.1242/jcs.002527 (doi:10.1242/jcs.002527) - DOI - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

