An E-cadherin-mediated hitchhiking mechanism for C. elegans germ cell internalization during gastrulation
- PMID: 22675206
- PMCID: PMC3383229
- DOI: 10.1242/dev.079863
An E-cadherin-mediated hitchhiking mechanism for C. elegans germ cell internalization during gastrulation
Abstract
Gastrulation movements place endodermal precursors, mesodermal precursors and primordial germ cells (PGCs) into the interior of the embryo. Somatic cell gastrulation movements are regulated by transcription factors that also control cell fate, coupling cell identity and position. By contrast, PGCs in many species are transcriptionally quiescent, suggesting that they might use alternative gastrulation strategies. Here, we show that C. elegans PGCs internalize by attaching to internal endodermal cells, which undergo morphogenetic movements that pull the PGCs into the embryo. We show that PGCs enrich HMR-1/E-cadherin at their surfaces to stick to endoderm. HMR-1 expression in PGCs is necessary and sufficient to ensure internalization, suggesting that HMR-1 can promote PGC-endoderm adhesion through a mechanism other than homotypic trans interactions between the two cell groups. Finally, we demonstrate that the hmr-1 3' untranslated region promotes increased HMR-1 translation in PGCs. Our findings reveal that quiescent PGCs employ a post-transcriptionally regulated hitchhiking mechanism to internalize during gastrulation, and demonstrate a morphogenetic role for the conserved association of PGCs with the endoderm.
Figures

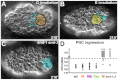
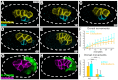
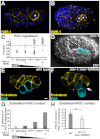



References
-
- Borghi N., Nelson W. J. (2009). Intercellular adhesion in morphogenesis: molecular and biochysical considerations. Curr. Top. Dev. Biol. 89, 1-32 - PubMed
-
- Broadbent I. D., Pettitt J. (2002). The C. elegans hmr-1 gene can encode a neuronal classic cadherin involved in the regulation of axon fasciculation. Curr. Biol. 12, 59-63 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

