On the mechanism of action of SJ-172550 in inhibiting the interaction of MDM4 and p53
- PMID: 22675482
- PMCID: PMC3366986
- DOI: 10.1371/journal.pone.0037518
On the mechanism of action of SJ-172550 in inhibiting the interaction of MDM4 and p53
Abstract
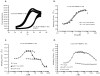
SJ-172550 (1) was previously discovered in a biochemical high throughput screen for inhibitors of the interaction of MDMX and p53 and characterized as a reversible inhibitor (J. Biol. Chem. 2010; 285:10786). Further study of the biochemical mode of action of 1 has shown that it acts through a complicated mechanism in which the compound forms a covalent but reversible complex with MDMX and locks MDMX into a conformation that is unable to bind p53. The relative stability of this complex is influenced by many factors including the reducing potential of the media, the presence of aggregates, and other factors that influence the conformational stability of the protein. This complex mechanism of action hinders the further development of compound 1 as a selective MDMX inhibitor.
Conflict of interest statement
Figures




References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
-
- Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7:979–987. - PubMed
-
- Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J Cell Sci. 2007;120:371–378. - PubMed
-
- Wynendaele J, Bohnke A, Leucci E, Nielsen SJ, Lambertz I, et al. An illegitimate microRNA target site within the 3' UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 2010;70:9641–9649. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

