Genes associated with MUC5AC expression in small airway epithelium of human smokers and non-smokers
- PMID: 22676183
- PMCID: PMC3443416
- DOI: 10.1186/1755-8794-5-21
Genes associated with MUC5AC expression in small airway epithelium of human smokers and non-smokers
Abstract
Background: Mucus hypersecretion contributes to the morbidity and mortality of smoking-related lung diseases, especially chronic obstructive pulmonary disease (COPD), which starts in the small airways. Despite progress in animal studies, the genes and their expression pattern involved in mucus production and secretion in human airway epithelium are not well understood. We hypothesized that comparison of the transcriptomes of the small airway epithelium of individuals that express high vs low levels of MUC5AC, the major macromolecular component of airway mucus, could be used as a probe to identify the genes related to human small airway mucus production/secretion.
Methods: Flexible bronchoscopy and brushing were used to obtain small airway epithelium (10th to 12th order bronchi) from healthy nonsmokers (n=60) and healthy smokers (n=72). Affymetrix HG-U133 plus 2.0 microarrays were used to assess gene expression. Massive parallel sequencing (RNA-Seq) was used to verify gene expression of small airway epithelium from 5 nonsmokers and 6 smokers.
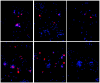
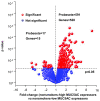
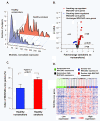
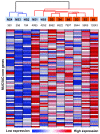
Results: MUC5AC expression varied 31-fold among the healthy nonsmokers. Genome-wide comparison between healthy nonsmokers (n = 60) grouped as "high MUC5AC expressors" vs "low MUC5AC expressors" identified 528 genes significantly up-regulated and 15 genes significantly down-regulated in the high vs low expressors. This strategy identified both mucus production and secretion related genes under control of a network composed of multiple transcription factors. Based on the literature, genes in the up-regulated list were used to identify a 73 "MUC5AC-associated core gene" list with 9 categories: mucus component; mucus-producing cell differentiation-related transcription factor; mucus-producing cell differentiation-related pathway or mediator; post-translational modification of mucin; vesicle transport; endoplasmic reticulum stress-related; secretory granule-associated; mucus secretion-related regulator and mucus hypersecretory-related ion channel. As a validation cohort, we assessed the MUC5AC-associated core gene list in the small airway epithelium of an independent set of healthy smokers (n = 72). There was up-regulation of MUC5AC in the small airway epithelium of smokers (2.3-fold, p < 10-8) associated with a coordinated up-regulation of MUC5AC-associated core gene expression pattern in the small airway epithelium of smokers (p < 0.01). Deep sequencing confirmed these observations.
Conclusion: The identification of the genes associated with increased airway mucin production in humans should be useful in understanding the pathogenesis of airway mucus hypersecretion and identifying therapeutic targets. AUTHOR SUMMARY: Mucus hypersecretion contributes to the morbidity and mortality of smoking-related lung diseases, especially chronic obstructive pulmonary disease (COPD), which starts in the small airways. Little is known about the gene networks associated with the synthesis and secretion of mucins in the human small airway epithelium. Taking advantage of the knowledge that MUC5AC is a major mucin secreted by the small airway epithelium, the expression of MUC5AC in small airway epithelium is highly regulated at the transcriptional level and our observation that healthy nonsmokers have variable numbers of MUC5AC+ secretory cells in the human small airway epithelium, we compared genome-wide gene expression of the small airway epithelium of high vs low MUC5AC expressors from 60 nonsmokers to identify the genes associated with MUC5AC expression. This novel strategy enabled identification of a 73 "MUC5AC-associated core gene" list with 9 categories, which control a series of processes from mucin biosynthesis to mucus secretion. The coordinated gene expression pattern of MUC5AC-associated core genes were corroborated in an independent cohort of 72 healthy smokers. Deep sequencing of small airway epithelium RNA confirmed these observations. This finding will be useful in identifying therapeutic targets to treat small airway mucus hypersecretion.
Figures

Similar articles
-
Smoking-mediated up-regulation of GAD67 expression in the human airway epithelium.Respir Res. 2010 Oct 29;11(1):150. doi: 10.1186/1465-9921-11-150. Respir Res. 2010. PMID: 21034448 Free PMC article.
-
Rho-kinase inhibitor fasudil reduces allergic airway inflammation and mucus hypersecretion by regulating STAT6 and NFκB.Clin Exp Allergy. 2015 Dec;45(12):1812-22. doi: 10.1111/cea.12606. Clin Exp Allergy. 2015. PMID: 26245530
-
Autophagy plays an essential role in cigarette smoke-induced expression of MUC5AC in airway epithelium.Am J Physiol Lung Cell Mol Physiol. 2016 Jun 1;310(11):L1042-52. doi: 10.1152/ajplung.00418.2015. Epub 2016 Apr 1. Am J Physiol Lung Cell Mol Physiol. 2016. PMID: 27036871
-
The Potential Role and Regulatory Mechanisms of MUC5AC in Chronic Obstructive Pulmonary Disease.Molecules. 2020 Sep 27;25(19):4437. doi: 10.3390/molecules25194437. Molecules. 2020. PMID: 32992527 Free PMC article. Review.
-
Mucins and Their Roles in Asthma.Immunol Rev. 2025 May;331(1):e70034. doi: 10.1111/imr.70034. Immunol Rev. 2025. PMID: 40305069 Review.
Cited by
-
Trypsin-Like Proteases and Their Role in Muco-Obstructive Lung Diseases.Int J Mol Sci. 2021 May 29;22(11):5817. doi: 10.3390/ijms22115817. Int J Mol Sci. 2021. PMID: 34072295 Free PMC article. Review.
-
The Role of Macrophages in the Development of Acute and Chronic Inflammatory Lung Diseases.Cells. 2021 Apr 14;10(4):897. doi: 10.3390/cells10040897. Cells. 2021. PMID: 33919784 Free PMC article. Review.
-
Update on the role of endoplasmic reticulum stress in asthma.Am J Transl Res. 2020 Apr 15;12(4):1168-1183. eCollection 2020. Am J Transl Res. 2020. PMID: 32355534 Free PMC article. Review.
-
Nasal gene expression differentiates COPD from controls and overlaps bronchial gene expression.Respir Res. 2017 Dec 21;18(1):213. doi: 10.1186/s12931-017-0696-5. Respir Res. 2017. PMID: 29268739 Free PMC article. Clinical Trial.
-
Tobacco Smoke Induces and Alters Immune Responses in the Lung Triggering Inflammation, Allergy, Asthma and Other Lung Diseases: A Mechanistic Review.Int J Environ Res Public Health. 2018 May 21;15(5):1033. doi: 10.3390/ijerph15051033. Int J Environ Res Public Health. 2018. PMID: 29883409 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

