Novel genetic linkage of rat Sp6 mutation to Amelogenesis imperfecta
- PMID: 22676574
- PMCID: PMC3464675
- DOI: 10.1186/1750-1172-7-34
Novel genetic linkage of rat Sp6 mutation to Amelogenesis imperfecta
Abstract
Background: Amelogenesis imperfecta (AI) is an inherited disorder characterized by abnormal formation of tooth enamel. Although several genes responsible for AI have been reported, not all causative genes for human AI have been identified to date. AMI rat has been reported as an autosomal recessive mutant with hypoplastic AI isolated from a colony of stroke-prone spontaneously hypertensive rat strain, but the causative gene has not yet been clarified. Through a genetic screen, we identified the causative gene of autosomal recessive AI in AMI and analyzed its role in amelogenesis.
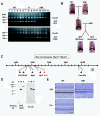
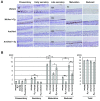
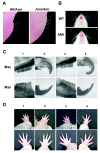
Methods: cDNA sequencing of possible AI-candidate genes so far identified using total RNA of day 6 AMI rat molars identified a novel responsible mutation in specificity protein 6 (Sp6). Genetic linkage analysis was performed between Sp6 and AI phenotype in AMI. To understand a role of SP6 in AI, we generated the transgenic rats harboring Sp6 transgene in AMI (Ami/Ami + Tg). Histological analyses were performed using the thin sections of control rats, AMI, and Ami/Ami + Tg incisors in maxillae, respectively.
Results: We found the novel genetic linkage between a 2-bp insertional mutation of Sp6 gene and the AI phenotype in AMI rats. The position of mutation was located in the coding region of Sp6, which caused frameshift mutation and disruption of the third zinc finger domain of SP6 with 11 cryptic amino acid residues and a stop codon. Transfection studies showed that the mutant protein can be translated and localized in the nucleus in the same manner as the wild-type SP6 protein. When we introduced the CMV promoter-driven wild-type Sp6 transgene into AMI rats, the SP6 protein was ectopically expressed in the maturation stage of ameloblasts associated with the extended maturation stage and the shortened reduced stage without any other phenotypical changes.
Conclusion: We propose the addition of Sp6 mutation as a new molecular diagnostic criterion for the autosomal recessive AI patients. Our findings expand the spectrum of genetic causes of autosomal recessive AI and sheds light on the molecular diagnosis for the classification of AI. Furthermore, tight regulation of the temporospatial expression of SP6 may have critical roles in completing amelogenesis.
Figures

References
-
- Witkop CJ Jr. Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: problems in classification. J Oral Pathol. 1988;17:547–553. - PubMed
-
- Cho SH, Seymen F, Lee KE, Lee SK, Kweon YS, Kim KJ, Jung SE, Song SJ, Yildirim M, Bayram M, Tuna EB, Gencay K, Kim JW. Novel FAM20A mutations in hypoplastic amelogenesis imperfecta. Hum Mutat. 2011;33:91–94. - PubMed
-
- O'Sullivan J, Bitu CC, Daly SB, Urquhart JE, Barron MJ, Bhaskar SS, Martelli-Junior H, dos Santos Neto PE, Mansilla MA, Murray JC, Coletta RD, Black GC, Dixon MJ. Whole-Exome sequencing identifies FAM20A mutations as a cause of amelogenesis imperfecta and gingival hyperplasia syndrome. Am J Hum Genet. 2011;88:616–620. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

