Probing the Xenopus laevis inner ear transcriptome for biological function
- PMID: 22676585
- PMCID: PMC3532188
- DOI: 10.1186/1471-2164-13-225
Probing the Xenopus laevis inner ear transcriptome for biological function
Abstract
Background: The senses of hearing and balance depend upon mechanoreception, a process that originates in the inner ear and shares features across species. Amphibians have been widely used for physiological studies of mechanotransduction by sensory hair cells. In contrast, much less is known of the genetic basis of auditory and vestibular function in this class of animals. Among amphibians, the genus Xenopus is a well-characterized genetic and developmental model that offers unique opportunities for inner ear research because of the amphibian capacity for tissue and organ regeneration. For these reasons, we implemented a functional genomics approach as a means to undertake a large-scale analysis of the Xenopus laevis inner ear transcriptome through microarray analysis.
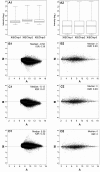
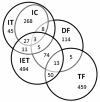
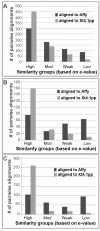
Results: Microarray analysis uncovered genes within the X. laevis inner ear transcriptome associated with inner ear function and impairment in other organisms, thereby supporting the inclusion of Xenopus in cross-species genetic studies of the inner ear. The use of gene categories (inner ear tissue; deafness; ion channels; ion transporters; transcription factors) facilitated the assignment of functional significance to probe set identifiers. We enhanced the biological relevance of our microarray data by using a variety of curation approaches to increase the annotation of the Affymetrix GeneChip(®) Xenopus laevis Genome array. In addition, annotation analysis revealed the prevalence of inner ear transcripts represented by probe set identifiers that lack functional characterization.
Conclusions: We identified an abundance of targets for genetic analysis of auditory and vestibular function. The orthologues to human genes with known inner ear function and the highly expressed transcripts that lack annotation are particularly interesting candidates for future analyses. We used informatics approaches to impart biologically relevant information to the Xenopus inner ear transcriptome, thereby addressing the impediment imposed by insufficient gene annotation. These findings heighten the relevance of Xenopus as a model organism for genetic investigations of inner ear organogenesis, morphogenesis, and regeneration.
Figures



References
-
- WHO. Deafness and hearing impairment. [ http://www.who.int/mediacentre/factsheets/fs300/en/]
-
- Statistics- Vestibular Disorders Association. Vertigo, Imbalance, Dizziness resources. [ http://www.vestibular.org/vestibular-disorders/statistics.php]
-
- Quick Statistics (NIDCD Health Information)
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

