Mutagenesis of RpoE-like sigma factor genes in Bdellovibrio reveals differential control of groEL and two groES genes
- PMID: 22676653
- PMCID: PMC3464611
- DOI: 10.1186/1471-2180-12-99
Mutagenesis of RpoE-like sigma factor genes in Bdellovibrio reveals differential control of groEL and two groES genes
Abstract
Background: Bdellovibrio bacteriovorus HD100 must regulate genes in response to a variety of environmental conditions as it enters, preys upon and leaves other bacteria, or grows axenically without prey. In addition to "housekeeping" sigma factors, its genome encodes several alternate sigma factors, including 2 Group IV-RpoE-like proteins, which may be involved in the complex regulation of its predatory lifestyle.
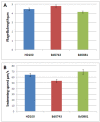
Results: We find that one sigma factor gene, bd3314, cannot be deleted from Bdellovibrio in either predatory or prey-independent growth states, and is therefore possibly essential, likely being an alternate sigma 70. Deletion of one of two Group IV-like sigma factor genes, bd0881, affects flagellar gene regulation and results in less efficient predation, although not due to motility changes; deletion of the second, bd0743, showed that it normally represses chaperone gene expression and intriguingly we find an alternative groES gene is expressed at timepoints in the predatory cycle where intensive protein synthesis at Bdellovibrio septation, prior to prey lysis, will be occurring.
Conclusions: We have taken the first step in understanding how alternate sigma factors regulate different processes in the predatory lifecycle of Bdellovibrio and discovered that alternate chaperones regulated by one of them are expressed at different stages of the lifecycle.
Figures




References
-
- Ruby EG. In: The Prokaryotes. 2. Schleifer KH, editor. Springer, New York; 1991. The genus Bdellovibrio.
-
- Shilo M, Bruff B. Lysis of Gram-negative bacteria by host-independent ectoparasitic Bdellovibrio bacteriovorus isolates. J Gen Microbiol. 1965;40:317–328. - PubMed
-
- Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A. et al.A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303(5658):689–692. - PubMed
-
- Heusipp G, Schmidt MA, Miller VL. Identification of rpoE and nadB as host responsive elements of Yersinia enterocolitica. FEMS Microbiol Lett. 2003;226(2):291–298. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

