Conformational dynamics of CYP3A4 demonstrate the important role of Arg212 coupled with the opening of ingress, egress and solvent channels to dehydrogenation of 4-hydroxy-tamoxifen
- PMID: 22677141
- PMCID: PMC3404218
- DOI: 10.1016/j.bbagen.2012.05.011
Conformational dynamics of CYP3A4 demonstrate the important role of Arg212 coupled with the opening of ingress, egress and solvent channels to dehydrogenation of 4-hydroxy-tamoxifen
Abstract
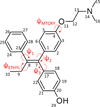
Background: Structure-based methods for P450 substrates are commonly used during drug development to identify sites of metabolism. However, docking studies using available X-ray structures for the major drug-metabolizing P450, CYP3A4, do not always identify binding modes supportive of the production of high-energy toxic metabolites. Minor pathways such as P450-catalyzed dehydrogenation have been experimentally shown to produce reactive products capable of forming biomolecular adducts which can lead to increased risk toxicities. 4-Hydroxy-tamoxifen (4OHT) is metabolized by CYP3A4 via competing hydroxylation and dehydrogenation reactions.
Methods: Ab initio gas-phase electronic structural characterization of 4OHT was used to develop a docking scoring scheme. Conformational sampling of CYP3A4 with molecular dynamics simulations along multiple trajectories were used to generate representative structures for docking studies using recently published heme parameters. A key predicted binding mode was tested experimentally using site-directed mutagenesis of CYP3A4 and liquid chromatography-mass spectroscopy analysis.
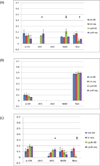
Results: Docking with MD-refined CYP3A4 structures incorporating hexa-coordinate heme parameters identifies a unique binding mode involving ARG212 and channel 4, unobserved in the starting PDB ID: 1TQN X-ray structure. The models supporting dehydrogenation are consistent with results from in vitro incubations.
General significance: Our models indicate that coupled structural contributions of the ingress, egress and solvent channels to the CYP3A4 active site geometries play key roles in the observed 4OHT binding modes. Thus adequate sampling of the conformational space of these drug-metabolizing promiscuous enzymes is important for substrates that may bind in malleable regions of the enzyme active-site.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures







Similar articles
-
Improved cytochrome P450 3A4 molecular models accurately predict the Phe215 requirement for raloxifene dehydrogenation selectivity.Biochemistry. 2010 Oct 19;49(41):9011-9. doi: 10.1021/bi101139q. Biochemistry. 2010. PMID: 20812728 Free PMC article.
-
Unraveling the Ligand-Binding Sites of CYP3A4 by Molecular Dynamics Simulations with Solvent Probes.J Chem Inf Model. 2024 Apr 22;64(8):3451-3464. doi: 10.1021/acs.jcim.4c00089. Epub 2024 Apr 9. J Chem Inf Model. 2024. PMID: 38593186
-
Computational Investigation of Ligand Binding to the Peripheral Site in CYP3A4: Conformational Dynamics and Inhibitor Discovery.J Chem Inf Model. 2017 Mar 27;57(3):616-626. doi: 10.1021/acs.jcim.7b00012. Epub 2017 Mar 2. J Chem Inf Model. 2017. PMID: 28221037
-
Challenges in assignment of allosteric effects in cytochrome P450-catalyzed substrate oxidations to structural dynamics in the hemoprotein architecture.J Inorg Biochem. 2017 Feb;167:100-115. doi: 10.1016/j.jinorgbio.2016.11.025. Epub 2016 Nov 25. J Inorg Biochem. 2017. PMID: 27919007 Review.
-
Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4.Curr Drug Metab. 2008 May;9(4):310-22. doi: 10.2174/138920008784220664. Curr Drug Metab. 2008. PMID: 18473749 Review.
Cited by
-
Homotropic Cooperativity of Midazolam Metabolism by Cytochrome P450 3A4: Insight from Computational Studies.J Chem Inf Model. 2021 May 24;61(5):2418-2426. doi: 10.1021/acs.jcim.1c00266. Epub 2021 Apr 22. J Chem Inf Model. 2021. PMID: 33884878 Free PMC article.
-
Elimination of tucatinib, a small molecule kinase inhibitor of HER2, is primarily governed by CYP2C8 enantioselective oxidation of gem-dimethyl.Cancer Chemother Pharmacol. 2022 Jun;89(6):737-750. doi: 10.1007/s00280-022-04429-z. Epub 2022 Apr 18. Cancer Chemother Pharmacol. 2022. PMID: 35435471
-
Pi-pi Stacking Mediated Cooperative Mechanism for Human Cytochrome P450 3A4.Molecules. 2015 Apr 24;20(5):7558-73. doi: 10.3390/molecules20057558. Molecules. 2015. PMID: 25919277 Free PMC article.
-
A multiscale approach to modelling drug metabolism by membrane-bound cytochrome P450 enzymes.PLoS Comput Biol. 2014 Jul 17;10(7):e1003714. doi: 10.1371/journal.pcbi.1003714. eCollection 2014 Jul. PLoS Comput Biol. 2014. PMID: 25033460 Free PMC article.
-
Molecular Dynamics Simulation Framework to Probe the Binding Hypothesis of CYP3A4 Inhibitors.Int J Mol Sci. 2019 Sep 10;20(18):4468. doi: 10.3390/ijms20184468. Int J Mol Sci. 2019. PMID: 31510073 Free PMC article.
References
-
- Furge LL, Guengerich FP. Cytochrome P450 enzymes in drug metabolism and chemical toxicology: An introduction. Biochemistry and Molecular Biology Education. 2006;34:66–74. - PubMed
-
- Ortiz de Montellano P, Voss J. In: Substrate Oxidation by Cytochrome P450 Enzymes Cytochrome P450. Ortiz de Montellano PR, editor. Springer US: 2005. pp. 183–245.
-
- Otyepka M, Skopalík J, Anzenbacherová E, Anzenbacher P. What common structural features and variations of mammalian P450s are known to date? Biochimica et Biophysica Acta (BBA) -General Subjects. 2007;1770:376–389. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

