Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation
- PMID: 22677446
- PMCID: PMC3408825
- DOI: 10.1016/j.tcb.2012.04.008
Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation
Abstract
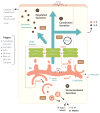
Autophagy is a cell biological process ubiquitous to all eukaryotic cells, often referred to as a catabolic, lysosomal degradative pathway. However, current studies in mammalian systems suggest that autophagy plays an unexpectedly broad biogenesis role in protein trafficking and secretion. Autophagy supports alternative trafficking pathways for delivery of integral membrane proteins to the plasma membrane and affects secretion, including the constitutive, regulated and unconventional secretion pathways. Autophagy-based unconventional secretion, termed here 'autosecretion', is one of the pathways enabling leaderless cytosolic proteins to exit the cell without entering the endoplasmic reticulum (ER)-to-Golgi secretory pathway. In this review, we discuss the emerging underlying mechanisms of how autophagy affects different facets of secretion. We also describe the physiological roles of autosecretory cargos that are often associated with inflammatory processes and also play a role in the formation of specialized tissues and in tissue remodeling, expanding the immediate sphere of influence of autophagy from the intracellular to the extracellular space.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


References
-
- Mizushima N, et al. The role of atg proteins in autophagosome formation. Annual review of cell and developmental biology. 2011;27:107–132. - PubMed
-
- Liu L, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–185. - PubMed
-
- Rubinsztein DC, et al. Autophagy and aging. Cell. 2011;146:682–695. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

