Heteroduplex formation, mismatch resolution, and genetic sectoring during homologous recombination in the hyperthermophilic archaeon sulfolobus acidocaldarius
- PMID: 22679441
- PMCID: PMC3367456
- DOI: 10.3389/fmicb.2012.00192
Heteroduplex formation, mismatch resolution, and genetic sectoring during homologous recombination in the hyperthermophilic archaeon sulfolobus acidocaldarius
Abstract
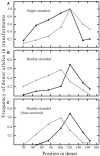
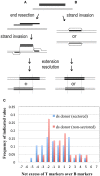
Hyperthermophilic archaea exhibit certain molecular-genetic features not seen in bacteria or eukaryotes, and their systems of homologous recombination (HR) remain largely unexplored in vivo. We transformed a Sulfolobus acidocaldariuspyrE mutant with short DNAs that contained multiple non-selected genetic markers within the pyrE gene. From 20 to 40% of the resulting colonies were found to contain two Pyr(+) clones with distinct sets of the non-selected markers. The dual-genotype colonies could not be attributed to multiple DNAs entering the cells, or to conjugation between transformed and non-transformed cells. These colonies thus appear to represent genetic sectoring in which regions of heteroduplex DNA formed and then segregated after partial resolution of inter-strand differences. Surprisingly, sectoring was also frequent in cells transformed with single-stranded DNAs. Oligonucleotides produced more sectored transformants when electroporated as single strands than as a duplex, although all forms of donor DNA (positive-strand, negative-strand, and duplex) produced a diversity of genotypes, despite the limited number of markers. The marker patterns in the recombinants indicate that S. acidocaldarius resolves individual mismatches through un-coordinated short-patch excision followed by re-filling of the resulting gap. The conversion events that occur during transformation by single-stranded DNA do not show the strand bias necessary for a system that corrects replication errors effectively; similar events also occur in pre-formed heteroduplex electroporated into the cells. Although numerous mechanistic details remain obscure, the results demonstrate that the HR system of S. acidocaldarius can generate remarkable genetic diversity from short intervals of moderately diverged DNAs.
Keywords: gene conversion; genetic transformation; linear DNA; mismatch repair.
Figures






References
LinkOut - more resources
Full Text Sources
Miscellaneous

