SUMO-activating SAE1 transcription is positively regulated by Myc
- PMID: 22679563
- PMCID: PMC3365806
SUMO-activating SAE1 transcription is positively regulated by Myc
Abstract
Myc protein plays a fundamental role in regulation of cell cycle, proliferation, differentiation and apoptosis by modulating the expression of a large number of targets. Here we report the transactivation ability of the human Myc protein to activate the SUMO-activating enzyme SAE1 transcription. We found that Myc activates SAE1 transcription via direct binding to canonical E-Boxes sequences located close to the SAE1 transcription start site. A recent report has highlighted the crucial role of the SAE gene expression in Myc mediated oncogenesis. Our study adds new insight in this context since we show here that Myc directly activates SAE1 transcription, suggesting that Myc oncogenic activity which depends on SAE1 is ensured by Myc itself through direct binding and transcriptional activation of SAE1 expression.
Keywords: Myc; SAE1; SUMOylation; transcription.
Figures
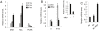


References
-
- Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. - PubMed
-
- Hay RT. SUMO. Mol Cell. 2005;18:1–7. - PubMed
-
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, Rao M, Yu P, Dominguez-Vidana R, Liang AC, Solimini NL, Bernardi RJ, Yu B, Hsu T, Golding I, Luo J, Osborne CK, Creighton CJ, Hilsenbeck SG, Schiff R, Shaw CA, Elledge SJ, Westbrook TF. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. - PMC - PubMed
LinkOut - more resources
Full Text Sources
