Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors
- PMID: 22681902
- PMCID: PMC3375611
- DOI: 10.1016/j.str.2012.04.010
Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors
Abstract
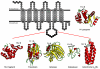
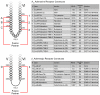
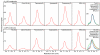
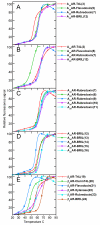
Structural studies of human G protein-coupled receptors (GPCRs) have recently been accelerated through the use of a fusion partner that was inserted into the third intracellular loop. Using chimeras of the human β(2)-adrenergic and human A(2A) adenosine receptors, we present the methodology and data for the initial selection of an expanded set of fusion partners for crystallizing GPCRs. In particular, use of the thermostabilized apocytochrome b(562)RIL as a fusion partner displays certain advantages over previously utilized fusion proteins, resulting in a significant improvement in stability and structure of GPCR-fusion constructs.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


References
-
- Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351–359. - PubMed
-
- Bjarnadottir TK, Gloriam DE, Hellstrand SH, Kristiansson H, Fredriksson R, Schioth HB. Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics. 2006;88:263–273. - PubMed
-
- Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallogr D Biol Crystallogr. 2004;60:1795–1807. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

