A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death
- PMID: 22683266
- PMCID: PMC3477315
- DOI: 10.1016/j.molcel.2012.05.004
A death effector domain chain DISC model reveals a crucial role for caspase-8 chain assembly in mediating apoptotic cell death
Abstract
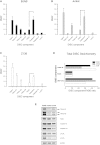
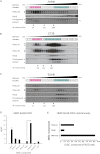
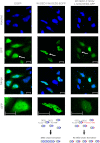
Formation of the death-inducing signaling complex (DISC) is a critical step in death receptor-mediated apoptosis, yet the mechanisms underlying assembly of this key multiprotein complex remain unclear. Using quantitative mass spectrometry, we have delineated the stoichiometry of the native TRAIL DISC. While current models suggest that core DISC components are present at a ratio of 1:1, our data indicate that FADD is substoichiometric relative to TRAIL-Rs or DED-only proteins; strikingly, there is up to 9-fold more caspase-8 than FADD in the DISC. Using structural modeling, we propose an alternative DISC model in which procaspase-8 molecules interact sequentially, via their DED domains, to form a caspase-activating chain. Mutating key interacting residues in procaspase-8 DED2 abrogates DED chain formation in cells and disrupts TRAIL/CD95 DISC-mediated procaspase-8 activation in a functional DISC reconstitution model. This provides direct experimental evidence for a DISC model in which DED chain assembly drives caspase-8 dimerization/activation, thereby triggering cell death.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures





References
-
- Berglund H., Olerenshaw D., Sankar A., Federwisch M., McDonald N.Q., Driscoll P.C. The three-dimensional solution structure and dynamic properties of the human FADD death domain. J. Mol. Biol. 2000;302:171–188. - PubMed
-
- Bertrand M.J., Vandenabeele P. The Ripoptosome: death decision in the cytosol. Mol. Cell. 2011;43:323–325. - PubMed
-
- Blondeau F., Ritter B., Allaire P.D., Wasiak S., Girard M., Hussain N.K., Angers A., Legendre-Guillemin V., Roy L., Boismenu D. Tandem MS analysis of brain clathrin-coated vesicles reveals their critical involvement in synaptic vesicle recycling. Proc. Natl. Acad. Sci. USA. 2004;101:3833–3838. - PMC - PubMed
-
- Boatright K.M., Renatus M., Scott F.L., Sperandio S., Shin H., Pedersen I.M., Ricci J.E., Edris W.A., Sutherlin D.P., Green D.R., Salvesen G.S. A unified model for apical caspase activation. Mol. Cell. 2003;11:529–541. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

