T cell antigen recognition at the cell membrane
- PMID: 22683645
- PMCID: PMC3403742
- DOI: 10.1016/j.molimm.2012.05.004
T cell antigen recognition at the cell membrane
Abstract
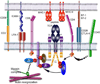
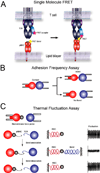
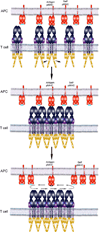
T cell antigen receptors (TCRs) on the surface of T cells bind specifically to particular peptide bound major histocompatibility complexes (pMHCs) presented on the surface of antigen presenting cells (APCs). This interaction is a key event in T cell antigen recognition and activation. Most studies have used surface plasmon resonance (SPR) to measure the in vitro binding kinetics of TCR-pMHC interactions in solution using purified proteins. However, these measurements are not physiologically precise, as both TCRs and pMHCs are membrane-associated molecules which are regulated by their cellular environments. Recently, single-molecule förster resonance energy transfer (FRET) and single-molecule mechanical assays were used to measure the in situ binding kinetics of TCR-pMHC interactions on the surface of live T cells. These studies have provided exciting insights into the biochemical basis of T cell antigen recognition and suggest that TCRs serially engage with a small number of antigens with very fast kinetics in order to maximize TCR signaling and sensitivity.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

References
-
- Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NR. T-cell-receptor affinity and thymocyte positive selection. Nature. 1996;381:616–620. - PubMed
-
- Alegre ML, Frauwirth KA, Thompson CB. T-cell regulation by CD28 and CTLA-4. Nat Rev Immunol. 2001;1:220–228. - PubMed
-
- Altschuh D, Dubs MC, Weiss E, Zeder-Lutz G, Van Regenmortel MH. Determination of kinetic constants for the interaction between a monoclonal antibody and peptides using surface plasmon resonance. Biochemistry. 1992;31:6298–6304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

