Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance
- PMID: 22683778
- PMCID: PMC3693569
- DOI: 10.1038/nm.2795
Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance
Abstract
Neoadjuvant chemotherapy (NAC) induces a pathological complete response (pCR) in ~30% of patients with breast cancer. However, many patients have residual cancer after chemotherapy, which correlates with a higher risk of metastatic recurrence and poorer outcome than those who achieve a pCR. We hypothesized that molecular profiling of tumors after NAC would identify genes associated with drug resistance. Digital transcript counting was used to profile surgically resected breast cancers after NAC. Low concentrations of dual specificity protein phosphatase 4 (DUSP4), an ERK phosphatase, correlated with high post-NAC tumor cell proliferation and with basal-like breast cancer (BLBC) status. BLBC had higher DUSP4 promoter methylation and gene expression patterns of Ras-ERK pathway activation relative to other breast cancer subtypes. DUSP4 overexpression increased chemotherapy-induced apoptosis, whereas DUSP4 depletion dampened the response to chemotherapy. Reduced DUSP4 expression in primary tumors after NAC was associated with treatment-refractory high Ki-67 scores and shorter recurrence-free survival. Finally, inhibition of mitogen-activated protein kinase kinase (MEK) synergized with docetaxel treatment in BLBC xenografts. Thus, DUSP4 downregulation activates the Ras-ERK pathway in BLBC, resulting in an attenuated response to anti-cancer chemotherapy.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
Figures


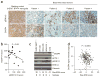

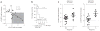

References
-
- Geiss GK, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- K99 CA142899/CA/NCI NIH HHS/United States
- P50 CA098258/CA/NCI NIH HHS/United States
- P50 CA098131/CA/NCI NIH HHS/United States
- R01 CA143126/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- P01 CA099031/CA/NCI NIH HHS/United States
- R00 CA142899/CA/NCI NIH HHS/United States
- P50CA98131/CA/NCI NIH HHS/United States
- P30CA68485/CA/NCI NIH HHS/United States
- BREAST CANCER NOW RESEARCH CENTRE/BBC_/Breast Cancer Now/United Kingdom
- P01CA099031/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

