Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia
- PMID: 22683780
- PMCID: PMC3438345
- DOI: 10.1038/nm.2819
Autocrine activation of the MET receptor tyrosine kinase in acute myeloid leukemia
Abstract
Although the treatment of acute myeloid leukemia (AML) has improved substantially in the past three decades, more than half of all patients develop disease that is refractory to intensive chemotherapy. Functional genomics approaches offer a means to discover specific molecules mediating the aberrant growth and survival of cancer cells. Thus, using a loss-of-function RNA interference genomic screen, we identified the aberrant expression of hepatocyte growth factor (HGF) as a crucial element in AML pathogenesis. We found HGF expression leading to autocrine activation of its receptor tyrosine kinase, MET, in nearly half of the AML cell lines and clinical samples we studied. Genetic depletion of HGF or MET potently inhibited the growth and survival of HGF-expressing AML cells. However, leukemic cells treated with the specific MET kinase inhibitor crizotinib developed resistance resulting from compensatory upregulation of HGF expression, leading to the restoration of MET signaling. In cases of AML where MET is coactivated with other tyrosine kinases, such as fibroblast growth factor receptor 1 (FGFR1), concomitant inhibition of FGFR1 and MET blocked this compensatory HGF upregulation, resulting in sustained logarithmic cell killing both in vitro and in xenograft models in vivo. Our results show a widespread dependence of AML cells on autocrine activation of MET, as well as the key role of compensatory upregulation of HGF expression in maintaining leukemogenic signaling by this receptor. We anticipate that these findings will lead to the design of additional strategies to block adaptive cellular responses that drive compensatory ligand expression as an essential component of the targeted inhibition of oncogenic receptors in human cancers.
Conflict of interest statement
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at
Figures
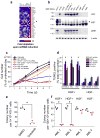
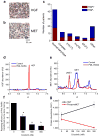
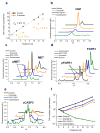
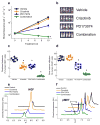
Comment in
-
The haves and the have nots.Nat Rev Cancer. 2012 Jul 24;12(8):505. doi: 10.1038/nrc3330. Nat Rev Cancer. 2012. PMID: 22825209 No abstract available.
-
Ligand-induced MET signaling as targetable codependence in acute myeloid leukemia.Haematologica. 2012 Aug;97(8):1118. doi: 10.3324/haematol.2012.073635. Haematologica. 2012. PMID: 22855844 Free PMC article. No abstract available.
References
-
- Grimwade D, et al. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001;98:1312–1320. - PubMed
-
- Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol. 2011;29:487–494. - PubMed
-
- Westbrook TF, Stegmeier F, Elledge SJ. Dissecting cancer pathways and vulnerabilities with RNAi. Cold Spring Harb Symp Quant Biol. 2005;70:435–444. - PubMed
-
- Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nat Methods. 2006;3:701–706. - PubMed
-
- Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

