The C-terminal repeating units of CsgB direct bacterial functional amyloid nucleation
- PMID: 22684146
- PMCID: PMC3423549
- DOI: 10.1016/j.jmb.2012.05.043
The C-terminal repeating units of CsgB direct bacterial functional amyloid nucleation
Abstract
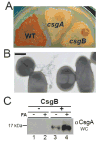
Curli are functional amyloids produced by enteric bacteria. The major curli fiber subunit, CsgA, self-assembles into an amyloid fiber in vitro. The minor curli subunit protein, CsgB, is required for CsgA polymerization on the cell surface. Both CsgA and CsgB are composed of five predicted β-strand-loop-β-strand-loop repeating units that feature conserved glutamine and asparagine residues. Because of this structural homology, we proposed that CsgB might form an amyloid template that initiates CsgA polymerization on the cell surface. To test this model, we purified wild-type CsgB and found that it self-assembled into amyloid fibers in vitro. Preformed CsgB fibers seeded CsgA polymerization as did soluble CsgB added to the surface of cells secreting soluble CsgA. To define the molecular basis of CsgB nucleation, we generated a series of mutants that removed each of the five repeating units. Each of these CsgB deletion mutants was capable of self-assembly in vitro. In vivo, membrane-localized mutants lacking the first, second, or third repeating units were able to convert CsgA into fibers. However, mutants missing either the fourth or fifth repeating units were unable to complement a csgB mutant. These mutant proteins were not localized to the outer membrane but were instead secreted into the extracellular milieu. Synthetic CsgB peptides corresponding to repeating units 1, 2, and 4 self-assembled into ordered amyloid polymers, while peptides corresponding to repeating units 3 and 5 did not, suggesting that there are redundant amyloidogenic domains in CsgB. Our results suggest a model where the rapid conversion of CsgB from unstructured protein to a β-sheet-rich amyloid template anchored to the cell surface is mediated by the C-terminal repeating units.
Published by Elsevier Ltd.
Figures



Similar articles
-
The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization.Proc Natl Acad Sci U S A. 2007 Jul 24;104(30):12494-9. doi: 10.1073/pnas.0703310104. Epub 2007 Jul 16. Proc Natl Acad Sci U S A. 2007. PMID: 17636121 Free PMC article.
-
Curli provide the template for understanding controlled amyloid propagation.Prion. 2008 Apr-Jun;2(2):57-60. doi: 10.4161/pri.2.2.6746. Epub 2008 Apr 5. Prion. 2008. PMID: 19098444 Free PMC article. Review.
-
Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis.Proc Natl Acad Sci U S A. 2010 Jan 5;107(1):163-8. doi: 10.1073/pnas.0908714107. Epub 2009 Dec 4. Proc Natl Acad Sci U S A. 2010. PMID: 19966296 Free PMC article.
-
Sequence determinants of bacterial amyloid formation.J Mol Biol. 2008 Jul 11;380(3):570-80. doi: 10.1016/j.jmb.2008.05.019. Epub 2008 May 17. J Mol Biol. 2008. PMID: 18565345 Free PMC article.
-
Curli biogenesis: order out of disorder.Biochim Biophys Acta. 2014 Aug;1843(8):1551-8. doi: 10.1016/j.bbamcr.2013.09.010. Epub 2013 Sep 27. Biochim Biophys Acta. 2014. PMID: 24080089 Free PMC article. Review.
Cited by
-
Plant Polyphenols Inhibit Functional Amyloid and Biofilm Formation in Pseudomonas Strains by Directing Monomers to Off-Pathway Oligomers.Biomolecules. 2019 Oct 26;9(11):659. doi: 10.3390/biom9110659. Biomolecules. 2019. PMID: 31717821 Free PMC article.
-
A semi high-throughput method for real-time monitoring of curli producing Salmonella biofilms on air-solid interfaces.Biofilm. 2021 Nov 13;3:100060. doi: 10.1016/j.bioflm.2021.100060. eCollection 2021 Dec. Biofilm. 2021. PMID: 34841245 Free PMC article.
-
Curli-Containing Enteric Biofilms Inside and Out: Matrix Composition, Immune Recognition, and Disease Implications.Microbiol Mol Biol Rev. 2018 Oct 10;82(4):e00028-18. doi: 10.1128/MMBR.00028-18. Print 2018 Dec. Microbiol Mol Biol Rev. 2018. PMID: 30305312 Free PMC article. Review.
-
Assembly of ordered DNA-curli fibril complexes during Salmonella biofilm formation correlates with strengths of the type I interferon and autoimmune responses.PLoS Pathog. 2022 Aug 16;18(8):e1010742. doi: 10.1371/journal.ppat.1010742. eCollection 2022 Aug. PLoS Pathog. 2022. PMID: 35972973 Free PMC article.
-
Electrostatic interactions mediate the nucleation and growth of a bacterial functional amyloid.Front Mol Biosci. 2023 Jan 12;10:1070521. doi: 10.3389/fmolb.2023.1070521. eCollection 2023. Front Mol Biosci. 2023. PMID: 36756360 Free PMC article.
References
-
- Cohen FE, Kelly JW. Therapeutic approaches to protein-misfolding diseases. Nature. 2003;426:905–9. - PubMed
-
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–66. - PubMed
-
- Jarrett JT, Lansbury PT., Jr Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 1993;73:1055–8. - PubMed
-
- Pedersen JS, Christensen G, Otzen DE. Modulation of S6 fibrillation by unfolding rates and gatekeeper residues. J Mol Biol. 2004;341:575–88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

