The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r
- PMID: 22684254
- PMCID: PMC3389517
- DOI: 10.1038/ncb2521
The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r
Abstract
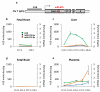
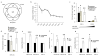
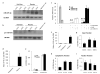
The H19 large intergenic non-coding RNA (lincRNA) is one of the most highly abundant and conserved transcripts in mammalian development, being expressed in both embryonic and extra-embryonic cell lineages, yet its physiological function is unknown. Here we show that miR-675, a microRNA (miRNA) embedded in H19's first exon, is expressed exclusively in the placenta from the gestational time point when placental growth normally ceases, and placentas that lack H19 continue to grow. Overexpression of miR-675 in a range of embryonic and extra-embryonic cell lines results in their reduced proliferation; targets of the miRNA are upregulated in the H19 null placenta, including the growth-promoting insulin-like growth factor 1 receptor (Igf1r) gene. Moreover, the excision of miR-675 from H19 is dynamically regulated by the stress-response RNA-binding protein HuR. These results suggest that H19's main physiological role is in limiting growth of the placenta before birth, by regulated processing of miR-675. The controlled release of miR-675 from H19 may also allow rapid inhibition of cell proliferation in response to cellular stress or oncogenic signals.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- 095645/WT_/Wellcome Trust/United Kingdom
- U01HL100407/HL/NHLBI NIH HHS/United States
- BB/H008071/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- G0801156/MRC_/Medical Research Council/United Kingdom
- BBS/E/B/0000S119/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

