Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs
- PMID: 22684503
- PMCID: PMC3439908
- DOI: 10.1093/nar/gks518
Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs
Abstract
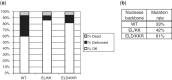
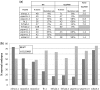
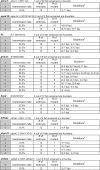
Transcription activator-like effector nucleases (TALENs) are powerful new research tools that enable targeted gene disruption in a wide variety of model organisms. Recent work has shown that TALENs can induce mutations in endogenous zebrafish genes, but to date only four genes have been altered, and larger-scale tests of the success rate, mutation efficiencies and germline transmission rates have not been described. Here, we constructed homodimeric TALENs to 10 different targets in various endogenous zebrafish genes and found that 7 nuclease pairs induced targeted indel mutations with high efficiencies ranging from 2 to 76%. We also tested obligate heterodimeric TALENs and found that these nucleases induce mutations with comparable or higher frequencies and have better toxicity profiles than their homodimeric counterparts. Importantly, mutations induced by both homodimeric and heterodimeric TALENs are passed efficiently through the germline, in some cases reaching 100% transmission. For one target gene sequence, we observed substantially reduced mutagenesis efficiency for a variant site bearing two mismatched nucleotides, raising the possibility that TALENs might be used to perform allele-specific gene disruption. Our results suggest that construction of one to two heterodimeric TALEN pairs for any given gene will, in most cases, enable researchers to rapidly generate knockout zebrafish.
Figures


References
-
- Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. - PubMed
-
- Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. - PubMed
-
- Sander JD, Yeh JR, Peterson RT, Joung JK. Engineering zinc finger nucleases for targeted mutagenesis of zebrafish. Methods Cell Biol. 2011;104:51–58. - PubMed
-
- Carroll D, Beumer KJ, Morton JJ, Bozas A, Trautman JK. Gene targeting in Drosophila and Caenorhabditis elegans with zinc-finger nucleases. Methods Mol. Biol. 2008;435:63–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

