Engineering domain fusion chimeras from I-OnuI family LAGLIDADG homing endonucleases
- PMID: 22684507
- PMCID: PMC3439895
- DOI: 10.1093/nar/gks502
Engineering domain fusion chimeras from I-OnuI family LAGLIDADG homing endonucleases
Abstract
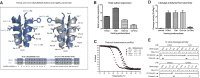
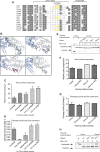
Although engineered LAGLIDADG homing endonucleases (LHEs) are finding increasing applications in biotechnology, their generation remains a challenging, industrial-scale process. As new single-chain LAGLIDADG nuclease scaffolds are identified, however, an alternative paradigm is emerging: identification of an LHE scaffold whose native cleavage site is a close match to a desired target sequence, followed by small-scale engineering to modestly refine recognition specificity. The application of this paradigm could be accelerated if methods were available for fusing N- and C-terminal domains from newly identified LHEs into chimeric enzymes with hybrid cleavage sites. Here we have analyzed the structural requirements for fusion of domains extracted from six single-chain I-OnuI family LHEs, spanning 40-70% amino acid identity. Our analyses demonstrate that both the LAGLIDADG helical interface residues and the linker peptide composition have important effects on the stability and activity of chimeric enzymes. Using a simple domain fusion method in which linker peptide residues predicted to contact their respective domains are retained, and in which limited variation is introduced into the LAGLIDADG helix and nearby interface residues, catalytically active enzymes were recoverable for ≈ 70% of domain chimeras. This method will be useful for creating large numbers of chimeric LHEs for genome engineering applications.
Figures



Similar articles
-
Engineering and flow-cytometric analysis of chimeric LAGLIDADG homing endonucleases from homologous I-OnuI-family enzymes.Methods Mol Biol. 2014;1123:191-221. doi: 10.1007/978-1-62703-968-0_14. Methods Mol Biol. 2014. PMID: 24510269 Free PMC article.
-
LAHEDES: the LAGLIDADG homing endonuclease database and engineering server.Nucleic Acids Res. 2012 Jul;40(Web Server issue):W110-6. doi: 10.1093/nar/gks365. Epub 2012 May 8. Nucleic Acids Res. 2012. PMID: 22570419 Free PMC article.
-
Expanding LAGLIDADG endonuclease scaffold diversity by rapidly surveying evolutionary sequence space.Nucleic Acids Res. 2012 Jun;40(11):4954-64. doi: 10.1093/nar/gkr1303. Epub 2012 Feb 14. Nucleic Acids Res. 2012. PMID: 22334611 Free PMC article.
-
Natural and engineered nicking endonucleases--from cleavage mechanism to engineering of strand-specificity.Nucleic Acids Res. 2011 Jan;39(1):1-18. doi: 10.1093/nar/gkq742. Epub 2010 Aug 30. Nucleic Acids Res. 2011. PMID: 20805246 Free PMC article. Review.
-
Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility.Nucleic Acids Res. 2001 Sep 15;29(18):3757-74. doi: 10.1093/nar/29.18.3757. Nucleic Acids Res. 2001. PMID: 11557808 Free PMC article. Review.
Cited by
-
Gene editing and its application for hematological diseases.Int J Hematol. 2016 Jul;104(1):18-28. doi: 10.1007/s12185-016-2017-z. Epub 2016 May 27. Int J Hematol. 2016. PMID: 27233509 Free PMC article. Review.
-
Homing endonucleases from mobile group I introns: discovery to genome engineering.Mob DNA. 2014 Mar 3;5(1):7. doi: 10.1186/1759-8753-5-7. Mob DNA. 2014. PMID: 24589358 Free PMC article.
-
DNA cleavage enzymes for treatment of persistent viral infections: recent advances and the pathway forward.Virology. 2014 Apr;454-455:353-61. doi: 10.1016/j.virol.2013.12.037. Epub 2014 Jan 31. Virology. 2014. PMID: 24485787 Free PMC article. Review.
-
Control of catalytic efficiency by a coevolving network of catalytic and noncatalytic residues.Proc Natl Acad Sci U S A. 2014 Jun 10;111(23):E2376-83. doi: 10.1073/pnas.1322352111. Epub 2014 May 27. Proc Natl Acad Sci U S A. 2014. PMID: 24912189 Free PMC article.
-
Efficient design of meganucleases using a machine learning approach.BMC Bioinformatics. 2014 Jun 17;15:191. doi: 10.1186/1471-2105-15-191. BMC Bioinformatics. 2014. PMID: 24934562 Free PMC article.
References
-
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. - PubMed
-
- Karran P. DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev. 2000;10:144–150. - PubMed
-
- McCammon JM, Amacher SL. Using zinc finger nucleases for efficient and heritable gene disruption in zebrafish. Methods Mol. Biol. 2010;649:281–298. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

