FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs
- PMID: 22684628
- PMCID: PMC3467035
- DOI: 10.1093/nar/gks542
FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs
Abstract
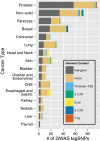
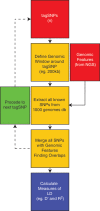
Single nucleotide polymorphisms (SNPs) are increasingly used to tag genetic loci associated with phenotypes such as risk of complex diseases. Technically, this is done genome-wide without prior restriction or knowledge of biological feasibility in scans referred to as genome-wide association studies (GWAS). Depending on the linkage disequilibrium (LD) structure at a particular locus, such tagSNPs may be surrogates for many thousands of other SNPs, and it is difficult to distinguish those that may play a functional role in the phenotype from those simply genetically linked. Because a large proportion of tagSNPs have been identified within non-coding regions of the genome, distinguishing functional from non-functional SNPs has been an even greater challenge. A strategy was recently proposed that prioritizes surrogate SNPs based on non-coding chromatin and epigenomic mapping techniques that have become feasible with the advent of massively parallel sequencing. Here, we introduce an R/Bioconductor software package that enables the identification of candidate functional SNPs by integrating information from tagSNP locations, lists of linked SNPs from the 1000 genomes project and locations of chromatin features which may have functional significance.
Availability: FunciSNP is available from Bioconductor (bioconductor.org).
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

