Topoisomerase assays
- PMID: 22684721
- PMCID: PMC3397423
- DOI: 10.1002/0471141755.ph0303s57
Topoisomerase assays
Abstract
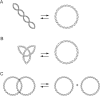
Topoisomerases are nuclear enzymes that play essential roles in DNA replication, transcription, chromosome segregation, and recombination. All cells have two major forms of topoisomerases: type I enzymes, which make single-stranded cuts in DNA, and type II enzymes, which cut and pass double-stranded DNA. DNA topoisomerases are important targets of approved and experimental anti-cancer agents. The protocols described in this unit are for assays used to assess new chemical entities for their ability to inhibit both forms of DNA topoisomerase. Included are an in vitro assay for topoisomerase I activity based on relaxation of supercoiled DNA, and an assay for topoisomerase II based on the decatenation of double-stranded DNA. The preparation of mammalian cell extracts for assaying topoisomerase activity is described, along with a protocol for an ICE assay to examine topoisomerase covalent complexes in vivo, and an assay for measuring DNA cleavage in vitro.
© 2012 by John Wiley & Sons, Inc.
Figures





References
-
- Albright LM, Slatko BE. Denaturing polyacrylamide gel electrophoresis. Curr Protoc Nucleic Acid Chem. 2001 Appendix 3:Appendix 3B. - PubMed
-
- Andoh T, Ishida R. Catalytic inhibitors of DNA topoisomerase II. Biochimica Et Biophysica Acta. 1998;1400:155–171. - PubMed
-
- Austin CA, Marsh KL, Wasserman RA, Willmore E, Sayer PJ, Wang JC, Fisher LM. Expression, domain structure, and enzymatic properties of an active recombinant human DNA topoisomerase II beta. J Biol Chem. 1995;270:15739–15746. - PubMed
-
- Austin CA, Sng JH, Patel S, Fisher LM. Novel HeLa topoisomerase II is the II beta isoform: complete coding sequence and homology with other type II topoisomerases. Biochim Biophys Acta. 1993;1172:283–291. - PubMed
-
- Bailly C. DNA relaxation and cleavage assays to study topoisomerase I inhibitors. Methods Enzymol. 2001;340:610–623. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

