Overlap of endocrine hormone expression in the mouse intestine revealed by transcriptional profiling and flow cytometry
- PMID: 22685263
- PMCID: PMC3440453
- DOI: 10.1210/en.2011-2170
Overlap of endocrine hormone expression in the mouse intestine revealed by transcriptional profiling and flow cytometry
Abstract
The intestine secretes a range of hormones with important local and distant actions, including the control of insulin secretion and appetite. A number of enteroendocrine cell types have been described, each characterized by a distinct hormonal signature, such as K-cells producing glucose-dependent insulinotropic polypeptide (GIP), L-cells producing glucagon-like peptide-1 (GLP-1), and I-cells producing cholecystokinin (CCK). To evaluate similarities between L-, K-, and other enteroendocrine cells, primary murine L- and K-cells, and pancreatic α- and β-cells, were purified and analyzed by flow cytometry and microarray-based transcriptomics. By microarray expression profiling, L cells from the upper small intestinal (SI) more closely resembled upper SI K-cells than colonic L-cells. Upper SI L-cell populations expressed message for hormones classically localized to different enteroendocrine cell types, including GIP, CCK, secretin, and neurotensin. By immunostaining and fluorescence-activated cell sorting analysis, most colonic L-cells contained GLP-1 and PeptideYY In the upper SI, most L-cells contained CCK, approximately 10% were GIP positive, and about 20% were PeptideYY positive. Upper SI K-cells exhibited approximately 10% overlap with GLP-1 and 6% overlap with somatostatin. Enteroendocrine-specific transcription factors were identified from the microarrays, of which very few differed between the enteroendocrine cell populations. Etv1, Prox1, and Pax4 were significantly enriched in L-cells vs. K cells by quantitative RT-PCR. In summary, our data indicate a strong overlap between upper SI L-, K-, and I-cells and suggest they may rather comprise a single cell type, within which individual cells exhibit a hormonal spectrum that may reflect factors such as location along the intestine and exposure to dietary nutrients.
Figures






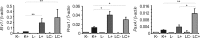
Comment in
-
A beautiful cell (or two or three?).Endocrinology. 2012 Jul;153(7):2945-8. doi: 10.1210/en.2012-1549. Endocrinology. 2012. PMID: 22730282 No abstract available.
References
-
- Holst JJ. 2007. The physiology of glucagon-like peptide 1. Physiol Rev 87:1409–1439 - PubMed
-
- Baggio LL, Drucker DJ. 2007. Biology of incretins: GLP-1 and GIP. Gastroenterology 132:2131–2157 - PubMed
-
- Dockray GJ. 2009. Cholecystokinin and gut-brain signalling. Regul Pept 155:6–10 - PubMed
-
- Karra E, Batterham RL. 2010. The role of gut hormones in the regulation of body weight and energy homeostasis. Mol Cell Endocrinol 316:120–128 - PubMed
-
- Ahrén B. 2011. The future of incretin-based therapy: novel avenues–novel targets. Diabetes Obes Metab 13(Suppl 1):158–166 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

