Type IV pilus assembly in Pseudomonas aeruginosa over a broad range of cyclic di-GMP concentrations
- PMID: 22685276
- PMCID: PMC3416225
- DOI: 10.1128/JB.00803-12
Type IV pilus assembly in Pseudomonas aeruginosa over a broad range of cyclic di-GMP concentrations
Abstract
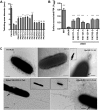
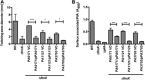
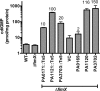
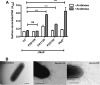
Pseudomonas aeruginosa is a Gram-negative, opportunistic pathogen that utilizes polar type IV pili (T4P) for twitching motility and adhesion in the environment and during infection. Pilus assembly requires FimX, a GGDEF/EAL domain protein that binds and hydrolyzes cyclic di-GMP (c-di-GMP). Bacteria lacking FimX are deficient in twitching motility and microcolony formation. We carried out an extragenic suppressor screen in PA103ΔfimX bacteria to identify additional regulators of pilus assembly. Multiple suppressor mutations were mapped to PA0171, PA1121 (yfiR), and PA3703 (wspF), three genes previously associated with small-colony-variant phenotypes. Multiple independent techniques confirmed that suppressors assembled functional surface pili, though at both polar and nonpolar sites. Whole-cell c-di-GMP levels were elevated in suppressor strains, in agreement with previous studies that had shown that the disrupted genes encoded negative regulators of diguanylate cyclases. Overexpression of the regulated diguanylate cyclases was sufficient to suppress the ΔfimX pilus assembly defect, as was overexpression of an unrelated diguanylate cyclase from Caulobacter crescentus. Furthermore, under natural conditions of high c-di-GMP, PA103ΔfimX formed robust biofilms that showed T4P staining and were structurally distinct from those formed by nonpiliated bacteria. These results are the first demonstration that P. aeruginosa assembles a surface organelle, type IV pili, over a broad range of c-di-GMP concentrations. Assembly of pili at low c-di-GMP concentrations requires a polarly localized c-di-GMP binding protein and phosphodiesterase, FimX; this requirement for FimX is bypassed at high c-di-GMP concentrations. Thus, P. aeruginosa can assemble the same surface organelle in distinct ways for motility or adhesion under very different environmental conditions.
Figures

Similar articles
-
Role of Cyclic Di-GMP and Exopolysaccharide in Type IV Pilus Dynamics.J Bacteriol. 2017 Mar 28;199(8):e00859-16. doi: 10.1128/JB.00859-16. Print 2017 Apr 15. J Bacteriol. 2017. PMID: 28167523 Free PMC article.
-
Interaction of the cyclic-di-GMP binding protein FimX and the Type 4 pilus assembly ATPase promotes pilus assembly.PLoS Pathog. 2017 Aug 30;13(8):e1006594. doi: 10.1371/journal.ppat.1006594. eCollection 2017 Aug. PLoS Pathog. 2017. PMID: 28854278 Free PMC article.
-
Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa.Mol Microbiol. 2006 May;60(4):1026-43. doi: 10.1111/j.1365-2958.2006.05156.x. Mol Microbiol. 2006. PMID: 16677312 Free PMC article.
-
c-di-GMP and its Effects on Biofilm Formation and Dispersion: a Pseudomonas Aeruginosa Review.Microbiol Spectr. 2015 Apr;3(2):MB-0003-2014. doi: 10.1128/microbiolspec.MB-0003-2014. Microbiol Spectr. 2015. PMID: 26104694 Free PMC article. Review.
-
Cyclic diguanylate signaling in Gram-positive bacteria.FEMS Microbiol Rev. 2016 Sep;40(5):753-73. doi: 10.1093/femsre/fuw013. Epub 2016 Jun 26. FEMS Microbiol Rev. 2016. PMID: 27354347 Free PMC article. Review.
Cited by
-
Genome-Wide Detection of Predicted Non-coding RNAs Related to the Adhesion Process in Vibrio alginolyticus Using High-Throughput Sequencing.Front Microbiol. 2016 Apr 28;7:619. doi: 10.3389/fmicb.2016.00619. eCollection 2016. Front Microbiol. 2016. PMID: 27199948 Free PMC article.
-
c-di-GMP-related phenotypes are modulated by the interaction between a diguanylate cyclase and a polar hub protein.Sci Rep. 2020 Feb 20;10(1):3077. doi: 10.1038/s41598-020-59536-9. Sci Rep. 2020. PMID: 32080219 Free PMC article.
-
Building permits-control of type IV pilus assembly by PilB and its cofactors.J Bacteriol. 2024 Dec 19;206(12):e0035924. doi: 10.1128/jb.00359-24. Epub 2024 Nov 7. J Bacteriol. 2024. PMID: 39508682 Free PMC article. Review.
-
Two-component systems required for virulence in Pseudomonas aeruginosa.FEMS Microbiol Lett. 2017 Jun 15;364(11):fnx104. doi: 10.1093/femsle/fnx104. FEMS Microbiol Lett. 2017. PMID: 28510688 Free PMC article. Review.
-
Assessment of polymicrobial interactions in bacterial isolates from transfused platelet units associated with sepsis.Transfusion. 2022 Dec;62(12):2458-2463. doi: 10.1111/trf.17136. Epub 2022 Sep 30. Transfusion. 2022. PMID: 36178430 Free PMC article.
References
-
- Barken KB, et al. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10: 2331–2343 - PubMed
-
- Bradley DE. 1980. A function of Pseudomonas aeruginosa PAO1 polar pili: twitching motility. Can. J. Microbiol. 26: 146–154 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

