Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18
- PMID: 22685290
- PMCID: PMC3406714
- DOI: 10.1074/jbc.M112.342113
Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18
Abstract
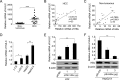
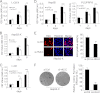
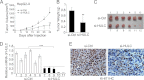
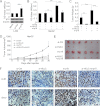
Long noncoding RNAs (lncRNAs) play crucial roles in human cancers. It has been reported that lncRNA highly up-regulated in liver cancer (HULC) is dramatically up-regulated in hepatocellular carcinoma (HCC). Hepatitis B virus X protein (HBx) contributes importantly to the development of HCC. However, the function of HULC in HCC mediated by HBx remains unclear. Here, we report that HULC is involved in HBx-mediated hepatocarcinogenesis. We found that the expression levels of HULC were positively correlated with those of HBx in clinical HCC tissues. Moreover, we revealed that HBx up-regulated HULC in human immortalized normal liver L-O2 cells and hepatoma HepG2 cells. Luciferase reporter gene assay and chromatin immunoprecipitation (ChIP) assay showed that HBx activated the HULC promoter via cAMP-responsive element-binding protein. We further demonstrated that HULC promoted cell proliferation by methyl thiazolyl tetrazolium, 5-ethynyl-2'-deoxyuridine, colony formation assay, and tumorigenicity assay. Next, we hypothesized that HULC might function through regulating a tumor suppressor gene p18 located near HULC in the same chromosome. We found that the mRNA levels of p18 were inversely correlated with those of HULC in the above clinical HCC specimens. Then, we validated that HULC down-regulated p18, which was involved in the HULC-enhanced cell proliferation in vitro and in vivo. Furthermore, we observed that knockdown of HULC could abolish the HBx-enhanced cell proliferation through up-regulating p18. Thus, we conclude that the up-regulated HULC by HBx promotes proliferation of hepatoma cells through suppressing p18. This finding provides new insight into the roles of lncRNAs in HBx-related hepatocarcinogenesis.
Figures


References
-
- El-Serag H. B., Rudolph K. L. (2007) Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132, 2557–2576 - PubMed
-
- Liu Q., Chen J., Liu L., Zhang J., Wang D., Ma L., He Y., Liu Y., Liu Z., Wu J. (2011) The X protein of hepatitis B virus inhibits apoptosis in hepatoma cells through enhancing the methionine adenosyltransferase 2A gene expression and reducing S-adenosylmethionine production. J. Biol. Chem. 286, 17168–17180 - PMC - PubMed
-
- Kim C. M., Koike K., Saito I., Miyamura T., Jay G. (1991) HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature 351, 317–320 - PubMed
-
- Lee S., Tarn C., Wang W. H., Chen S., Hullinger R. L., Andrisani O. M. (2002) Hepatitis B virus X protein differentially regulates cell cycle progression in X-transforming versus nontransforming hepatocyte (AML12) cell lines. J. Biol. Chem. 277, 8730–8740 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

