Protein dynamics and the diversity of an antibody response
- PMID: 22685303
- PMCID: PMC3411056
- DOI: 10.1074/jbc.M112.372698
Protein dynamics and the diversity of an antibody response
Abstract
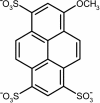
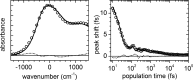
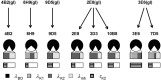
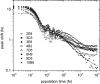
The immune system is remarkable in its ability to produce antibodies (Abs) with virtually any specificity from a limited repertoire of germ line precursors. Although the contribution of sequence diversity to this molecular recognition has been studied for decades, recent models suggest that protein dynamics may also broaden the range of targets recognized. To characterize the contribution of protein dynamics to immunological molecular recognition, we report the sequence, thermodynamic, and time-resolved spectroscopic characterization of a panel of eight Abs elicited to the chromophoric antigen 8-methoxypyrene-1,3,6-trisulfonate (MPTS). Based on the sequence data, three of the Abs arose from unique germ line Abs, whereas the remaining five comprise two sets of siblings that arose by somatic mutation of a common precursor. The thermodynamic data indicate that the Abs recognize MPTS via a variety of mechanisms. Although the spectroscopic data reveal small differences in protein dynamics, the anti-MPTS Abs generally show similar levels of flexibility and conformational heterogeneity, possibly representing the convergent evolution of the dynamics necessary for function. However, one Ab is significantly more rigid and conformationally homogeneous than the others, including a sibling Ab from which it differs by only five somatic mutations. This example of divergent evolution demonstrates that point mutations are capable of fixing significant differences in protein dynamics. The results provide unique insight into how high affinity Abs may be produced that bind virtually any target and possibly, from a more general perspective, how new protein functions are evolved.
Figures


Similar articles
-
Adaptive mutations alter antibody structure and dynamics during affinity maturation.Biochemistry. 2015 Mar 24;54(11):2085-93. doi: 10.1021/bi501417q. Epub 2015 Mar 10. Biochemistry. 2015. PMID: 25756188 Free PMC article.
-
Structure and Dynamics of Stacking Interactions in an Antibody Binding Site.Biochemistry. 2019 Jul 9;58(27):2987-2995. doi: 10.1021/acs.biochem.9b00119. Epub 2019 Jun 19. Biochemistry. 2019. PMID: 31243995 Free PMC article.
-
Exploring the energy landscape of antibody-antigen complexes: protein dynamics, flexibility, and molecular recognition.Biochemistry. 2008 Jul 8;47(27):7237-47. doi: 10.1021/bi800374q. Epub 2008 Jun 13. Biochemistry. 2008. PMID: 18549243
-
On the benefits of sin: can greater understanding of the 1F7-idiotypic repertoire freeze enhance HIV vaccine development?Hum Vaccin Immunother. 2013 Jul;9(7):1532-8. doi: 10.4161/hv.24460. Epub 2013 Apr 9. Hum Vaccin Immunother. 2013. PMID: 23571168 Review.
-
The biological origin of antibody diversity.Annu Rev Biochem. 1976;45:467-500. doi: 10.1146/annurev.bi.45.070176.002343. Annu Rev Biochem. 1976. PMID: 60909 Review.
Cited by
-
Targeting Stereotyped B Cell Receptors from Chronic Lymphocytic Leukemia Patients with Synthetic Antigen Surrogates.J Biol Chem. 2016 Apr 1;291(14):7558-70. doi: 10.1074/jbc.M115.701656. Epub 2016 Feb 5. J Biol Chem. 2016. PMID: 26851280 Free PMC article.
-
Probing the Kinetic and Thermodynamic Fingerprints of Anti-EGF Nanobodies by Surface Plasmon Resonance.Pharmaceuticals (Basel). 2020 Jun 26;13(6):134. doi: 10.3390/ph13060134. Pharmaceuticals (Basel). 2020. PMID: 32604841 Free PMC article.
-
Conformational Plasticity in Broadly Neutralizing HIV-1 Antibodies Triggers Polyreactivity.Cell Rep. 2018 May 29;23(9):2568-2581. doi: 10.1016/j.celrep.2018.04.101. Cell Rep. 2018. PMID: 29847789 Free PMC article.
-
Adaptive mutations alter antibody structure and dynamics during affinity maturation.Biochemistry. 2015 Mar 24;54(11):2085-93. doi: 10.1021/bi501417q. Epub 2015 Mar 10. Biochemistry. 2015. PMID: 25756188 Free PMC article.
-
Effective harmonic potentials: insights into the internal cooperativity and sequence-specificity of protein dynamics.PLoS Comput Biol. 2013;9(8):e1003209. doi: 10.1371/journal.pcbi.1003209. Epub 2013 Aug 29. PLoS Comput Biol. 2013. PMID: 24009495 Free PMC article.
References
-
- Gao J., Byun K. L., Kluger R. (2004) Catalysis by enzyme conformational change. Top. Curr. Chem. 238, 113–136
-
- Eisenmesser E. Z., Bosco D. A., Akke M., Kern D. (2002) Enzyme dynamics during catalysis. Science 295, 1520–1523 - PubMed
-
- Daniel R. M., Dunn R. V., Finney J. L., Smith J. C. (2003) The role of dynamics in enzyme activity. Annu. Rev. Biophys. Biomol. Struct. 32, 69–92 - PubMed
-
- Villa J., Warshel A. (2001) Energetics and dynamics of enzymatic reactions. J. Phys. Chem. B 105, 7887–7907
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

