Advances in high-resolution imaging--techniques for three-dimensional imaging of cellular structures
- PMID: 22685332
- PMCID: PMC3706075
- DOI: 10.1242/jcs.090027
Advances in high-resolution imaging--techniques for three-dimensional imaging of cellular structures
Abstract
A fundamental goal in biology is to determine how cellular organization is coupled to function. To achieve this goal, a better understanding of organelle composition and structure is needed. Although visualization of cellular organelles using fluorescence or electron microscopy (EM) has become a common tool for the cell biologist, recent advances are providing a clearer picture of the cell than ever before. In particular, advanced light-microscopy techniques are achieving resolutions below the diffraction limit and EM tomography provides high-resolution three-dimensional (3D) images of cellular structures. The ability to perform both fluorescence and electron microscopy on the same sample (correlative light and electron microscopy, CLEM) makes it possible to identify where a fluorescently labeled protein is located with respect to organelle structures visualized by EM. Here, we review the current state of the art in 3D biological imaging techniques with a focus on recent advances in electron microscopy and fluorescence super-resolution techniques.
Figures

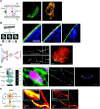
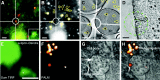
References
-
- Agronskaia A. V., Valentijn J. A., van Driel L. F., Schneijdenberg C. T., Humbel B. M., van Bergen en Henegouwen P. M., Verkleij A. J., Koster A. J., Gerritsen H. C. (2008). Integrated fluorescence and transmission electron microscopy. J. Struct. Biol. 164, 183-189 - PubMed
-
- Aquino D., Schönle A., Geisler C., Middendorff C. V., Wurm C. A., Okamura Y., Lang T., Hell S. W., Egner A. (2011). Two-color nanoscopy of three-dimensional volumes by 4Pi detection of stochastically switched fluorophores. Nat. Methods 8, 353-359 - PubMed
-
- Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J., Hess H. F. (2006). Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642-1645 - PubMed
-
- Bos E., SantAnna C., Gnaegi H., Pinto R. F., Ravelli R. B., Koster A. J., de Souza W., Peters P. J. (2011). A new approach to improve the quality of ultrathin cryo-sections; its use for immunogold EM and correlative electron cryo-tomography. J. Struct. Biol. 175, 62-72 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

