Pathogen-mediated proteolysis of the cell death regulator RIPK1 and the host defense modulator RIPK2 in human aortic endothelial cells
- PMID: 22685397
- PMCID: PMC3369954
- DOI: 10.1371/journal.ppat.1002723
Pathogen-mediated proteolysis of the cell death regulator RIPK1 and the host defense modulator RIPK2 in human aortic endothelial cells
Abstract
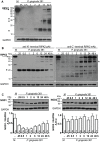
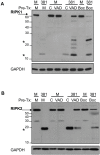
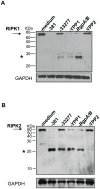
Porphyromonas gingivalis is the primary etiologic agent of periodontal disease that is associated with other human chronic inflammatory diseases, including atherosclerosis. The ability of P. gingivalis to invade and persist within human aortic endothelial cells (HAEC) has been postulated to contribute to a low to moderate chronic state of inflammation, although how this is specifically achieved has not been well defined. In this study, we demonstrate that P. gingivalis infection of HAEC resulted in the rapid cleavage of receptor interacting protein 1 (RIPK1), a mediator of tumor necrosis factor (TNF) receptor-1 (TNF-R1)-induced cell activation or death, and RIPK2, a key mediator of both innate immune signaling and adaptive immunity. The cleavage of RIPK1 or RIPK2 was not observed in cells treated with apoptotic stimuli, or cells stimulated with agonists to TNF-R1, nucleotide oligomerization domain receptor 1(NOD1), NOD2, Toll-like receptor 2 (TLR2) or TLR4. P. gingivalis-induced cleavage of RIPK1 and RIPK2 was inhibited in the presence of a lysine-specific gingipain (Kgp) inhibitor. RIPK1 and RIPK2 cleavage was not observed in HAEC treated with an isogenic mutant deficient in the lysine-specific gingipain, confirming a role for Kgp in the cleavage of RIPK1 and RIPK2. Similar proteolysis of poly (ADP-ribose) polymerase (PARP) was observed. We also demonstrated direct proteolysis of RIPK2 by P. gingivalis in a cell-free system which was abrogated in the presence of a Kgp-specific protease inhibitor. Our studies thus reveal an important role for pathogen-mediated modification of cellular kinases as a potential strategy for bacterial persistence within target host cells, which is associated with low-grade chronic inflammation, a hallmark of pathogen-mediated chronic inflammatory disorders.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
-
- Inohara N, del Peso L, Koseki T, Chen S, Nunez G. RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J Biol Chem. 1998;273:12296–12300. - PubMed
-
- Chin AI, Dempsey PW, Bruhn K, Miller JF, Xu Y. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature. 2002;416:190–194. - PubMed
-
- Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. - PubMed
-
- Zhang N, Chen Y, Jiang R, Li E, Chen X. PARP and RIP 1 are required for autophagy induced by 11′-deoxyverticillin A, which precedes caspase-dependent apoptosis. Autophagy. 2011;7:598–612. - PubMed
-
- Travassos LH, Carneiro LA, Girardin SE, Boneca IG, Lemos R. Nod1 participates in the innate immune response to Pseudomonas aeruginosa. J Biol Chem. 2005;280:36714–36718. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

