HIV-1 antiretroviral resistance: scientific principles and clinical applications
- PMID: 22686620
- PMCID: PMC3689909
- DOI: 10.2165/11633630-000000000-00000
HIV-1 antiretroviral resistance: scientific principles and clinical applications
Abstract
The efficacy of an antiretroviral (ARV) treatment regimen depends on the activity of the regimen's individual ARV drugs and the number of HIV-1 mutations required for the development of resistance to each ARV - the genetic barrier to resistance. ARV resistance impairs the response to therapy in patients with transmitted resistance, unsuccessful initial ARV therapy and multiple virological failures. Genotypic resistance testing is used to identify transmitted drug resistance, provide insight into the reasons for virological failure in treated patients, and help guide second-line and salvage therapies. In patients with transmitted drug resistance, the virological response to a regimen selected on the basis of standard genotypic testing approaches the responses observed in patients with wild-type viruses. However, because such patients are at a higher risk of harbouring minority drug-resistant variants, initial ARV therapy in this population should contain a boosted protease inhibitor (PI) - the drug class with the highest genetic barrier to resistance. In patients receiving an initial ARV regimen with a high genetic barrier to resistance, the most common reasons for virological failure are nonadherence and, potentially, pharmacokinetic factors or minority transmitted drug-resistant variants. Among patients in whom first-line ARVs have failed, the patterns of drug-resistance mutations and cross-resistance are often predictable. However, the extent of drug resistance correlates with the duration of uncontrolled virological replication. Second-line therapy should include the continued use of a dual nucleoside/nucleotide reverse transcriptase inhibitor (NRTI)-containing backbone, together with a change in the non-NRTI component, most often to an ARV belonging to a new drug class. The number of available fully active ARVs is often diminished with each successive treatment failure. Therefore, a salvage regimen is likely to be more complicated in that it may require multiple ARVs with partial residual activity and compromised genetic barriers of resistance to attain complete virological suppression. A thorough examination of the patient's ARV history and prior resistance tests should be performed because genotypic and/or phenotypic susceptibility testing is often not sufficient to identify drug-resistant variants that emerged during past therapies and may still pose a threat to a new regimen. Phenotypic testing is also often helpful in this subset of patients. ARVs used for salvage therapy can be placed into the following hierarchy: (i) ARVs belonging to a previously unused drug class; (ii) ARVs belonging to a previously used drug class that maintain significant residual antiviral activity; (iii) NRTI combinations, as these often appear to retain in vivo virological activity, even in the presence of reduced in vitro NRTI susceptibility; and rarely (iv) ARVs associated with previous virological failure and drug resistance that appear to have possibly regained their activity as a result of viral reversion to wild type. Understanding the basic principles of HIV drug resistance is helpful in guiding individual clinical decisions and the development of ARV treatment guidelines.
Figures



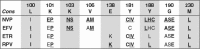
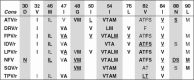



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

