In vitro capping and transcription of rhabdoviruses
- PMID: 22687619
- PMCID: PMC3449051
- DOI: 10.1016/j.ymeth.2012.05.013
In vitro capping and transcription of rhabdoviruses
Abstract
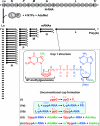
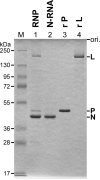
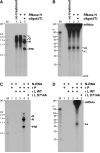
The RNA-dependent RNA polymerase L protein of vesicular stomatitis virus (VSV), a prototypic nonsegmented negative strand (NNS) RNA virus classified into the Rhabdoviridae family, has been used to investigate the fundamental molecular mechanisms of NNS RNA viral mRNA synthesis and processing. In vitro studies on mRNA cap formation with the VSV L protein eventually led to the discovery of the unconventional mRNA capping pathway catalyzed by the guanosine 5'-triphosphatase and RNA:GDP polyribonucleotidyltransferase (PRNTase) activities. The PRNTase activity is a novel enzymatic activity, which transfers 5'-monophosphorylated (p-) RNA from 5'-triphosphorylated (ppp-) RNA to GDP to form 5'-capped RNA (GpppRNA) in a viral mRNA-start sequence-dependent manner. This unconventional capping (pRNA transfer) reaction with PRNTase can be experimentally distinguished from the conventional capping (GMP transfer) reaction with eukaryotic GTP:RNA guanylyltransferase (GTase) on the basis of the following differences in their substrate specificity for the cap formation: PRNTase uses GDP and pppRNA, but not ppRNA, whereas GTase employs GTP, but not GDP, and ppRNA. The pRNA transfer reaction with PRNTase proceeds through a covalent enzyme-pRNA intermediate with a phosphoamide bond. Hence, to prove the PRNTase activity, it is necessary to demonstrate the following consecutive steps separately: (1) the enzyme forms a covalent enzyme-pRNA intermediate, and (2) the intermediate transfers pRNA to GDP. This article describes the methods for in vitro transcription and capping with the recombinant VSV L protein, which permit detailed characterization of its enzymatic reactions and mapping of active sites of its enzymatic domains. It is expected that these systems are adaptable to rhabdoviruses and, by extension, other NNS RNA viruses belonging to different families.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
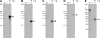




References
-
- Lyles DS, Rupprecht CE. In: Fields Virology Lippincott. Knipe DM, Howley PM, editors. Williams & Wilkins; Philadelphia: 2007. pp. 1363–1408.
-
- Poch O, Blumberg BM, Bougueleret L, Tordo N. J. Gen. Virol. 1990;71:1153–1162. - PubMed
-
- Volchkov VE, Volchkova VA, Chepurnov AA, Blinov VM, Dolnik O, Netesov SV, Feldmann H. J. Gen. Virol. 1999;80:355–362. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

