Metagenomic microbial community profiling using unique clade-specific marker genes
- PMID: 22688413
- PMCID: PMC3443552
- DOI: 10.1038/nmeth.2066
Metagenomic microbial community profiling using unique clade-specific marker genes
Abstract
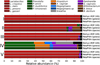
Metagenomic shotgun sequencing data can identify microbes populating a microbial community and their proportions, but existing taxonomic profiling methods are inefficient for increasingly large data sets. We present an approach that uses clade-specific marker genes to unambiguously assign reads to microbial clades more accurately and >50× faster than current approaches. We validated our metagenomic phylogenetic analysis tool, MetaPhlAn, on terabases of short reads and provide the largest metagenomic profiling to date of the human gut. It can be accessed at http://huttenhower.sph.harvard.edu/metaphlan/.
Figures
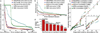

Comment in
-
High-speed microbial community profiling.Nat Methods. 2012 Jun 10;9(8):793-4. doi: 10.1038/nmeth.2080. Nat Methods. 2012. PMID: 22688412 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

