Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin
- PMID: 22688965
- PMCID: PMC3388787
- DOI: 10.1038/embor.2012.78
Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin
Erratum in
- EMBO Rep. 2012 Dec;13(12):1152
Abstract
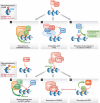
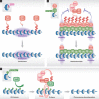
The organization of eukaryotic chromosomes into transcriptionally active euchromatin and repressed heterochromatin requires mechanisms that establish, maintain and distinguish these canonical chromatin domains. Post-translational modifications are fundamental in these processes. Monoubiquitylation of histones was discovered more than three decades ago, but its precise function has been enigmatic until recently. It is now appreciated that the spectrum of chromatin ubiquitylation is not restricted to monoubiquitylation of histones, but includes degradatory ubiquitylation of histones, histone-modifying enzymes and non-histone chromatin factors. These occur in a spatially and temporally controlled manner. In this review, we summarize our understanding of these mechanisms with a particular emphasis on how ubiquitylation shapes the physical landscape of chromatin.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Muratani M, Tansey WP (2003) How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol 4: 192–201 - PubMed
-
- Moldovan G-L, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129: 665–679 - PubMed
-
- Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 11: 261–266 - PubMed

