Peptide bicycles that inhibit the Grb2 SH2 domain
- PMID: 22689355
- PMCID: PMC3516885
- DOI: 10.1002/cbic.201200175
Peptide bicycles that inhibit the Grb2 SH2 domain
Abstract
Developing short peptides into useful probes and therapeutic leads remains a difficult challenge. Structural rigidification is a proven method for improving the properties of short peptides. In this work, we report a strategy for stabilizing peptide macrocycles by introducing side-chain-to-side-chain staples to produce peptide bicycles with higher affinity, selectivity, and resistance to degradation. We have applied this strategy to G1, an 11-residue peptide macrocycle that binds the Src homology 2 (SH2) domain of growth-factor-bound protein 2 (Grb2). Several homodetic peptide bicycles were synthesized entirely on-resin with high yields. Two rounds of iterative design produced peptide bicycle BC1, which is 60 times more potent than G1 and 200 times more selective. Moreover, BC1 is completely intact after 24 hours in buffered human serum, conditions under which G1 is completely degraded. Our peptide-bicycle approach holds promise for the development of selective inhibitors of SH2 domains and other phosophotyrosine (pTyr)-binding proteins, as well as inhibitors of many other protein-protein interactions.
Copyright © 2012 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures


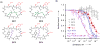



Similar articles
-
Structural basis for a non-phosphorus-containing cyclic peptide binding to Grb2-SH2 domain with high affinity.Biochem Biophys Res Commun. 2003 Aug 8;307(4):1038-44. doi: 10.1016/s0006-291x(03)01291-9. Biochem Biophys Res Commun. 2003. PMID: 12878216
-
Utilization of a beta-aminophosphotyrosyl mimetic in the design and synthesis of macrocyclic Grb2 SH2 domain-binding peptides.J Med Chem. 2003 Jun 19;46(13):2621-30. doi: 10.1021/jm030049q. J Med Chem. 2003. PMID: 12801226
-
Discovery of a novel nonphosphorylated pentapeptide motif displaying high affinity for Grb2-SH2 domain by the utilization of 3'-substituted tyrosine derivatives.J Med Chem. 2006 Mar 9;49(5):1585-96. doi: 10.1021/jm050910x. J Med Chem. 2006. PMID: 16509576
-
The Role of Grb2 in Cancer and Peptides as Grb2 Antagonists.Protein Pept Lett. 2018 Feb 8;24(12):1084-1095. doi: 10.2174/0929866525666171123213148. Protein Pept Lett. 2018. PMID: 29173143 Review.
-
Grb2 SH2 domain-binding peptide analogs as potential anticancer agents.Biopolymers. 2003;71(2):132-40. doi: 10.1002/bip.10396. Biopolymers. 2003. PMID: 12767115 Review.
Cited by
-
Probing SH2-domains using Inhibitor Affinity Purification (IAP).Proteome Sci. 2014 Jul 16;12:41. doi: 10.1186/1477-5956-12-41. eCollection 2014. Proteome Sci. 2014. PMID: 25067910 Free PMC article.
-
Rational Design of Peptide-Based Inhibitors Disrupting Protein-Protein Interactions.Front Chem. 2021 May 4;9:682675. doi: 10.3389/fchem.2021.682675. eCollection 2021. Front Chem. 2021. PMID: 34017824 Free PMC article. Review.
-
Targeting kinase signaling pathways with constrained peptide scaffolds.Pharmacol Ther. 2017 May;173:159-170. doi: 10.1016/j.pharmthera.2017.02.014. Epub 2017 Feb 7. Pharmacol Ther. 2017. PMID: 28185915 Free PMC article. Review.
-
Getting in shape: controlling peptide bioactivity and bioavailability using conformational constraints.ACS Chem Biol. 2013 Mar 15;8(3):488-499. doi: 10.1021/cb300515u. Epub 2012 Nov 30. ACS Chem Biol. 2013. PMID: 23170954 Free PMC article. Review.
-
SH2 Domains: Folding, Binding and Therapeutical Approaches.Int J Mol Sci. 2022 Dec 15;23(24):15944. doi: 10.3390/ijms232415944. Int J Mol Sci. 2022. PMID: 36555586 Free PMC article. Review.
References
-
- Kritzer JA. Nat Chem Biol. 2010;6:868–870. - PubMed
-
- Hruby VJ, Balse PM. Curr Med Chem. 2000;7:945–970. - PubMed
- Hruby VJ, Alobeidi F, Kazmierski W. Biochem J. 1990;268:249–262. - PMC - PubMed
- Pelton JT, Kazmierski W, Gulya K, Yamamura HI, Hruby VJ. J Med Chem. 1986;29:2370–2375. - PubMed
- Ovadia O, Greenberg S, Laufer B, Gilon C, Hoffman A, Kessler H. Exp Op Drug Disc. 2010;5:655–671. - PubMed
- Kessler H. Angew Chem Int Ed Engl. 1982;21:512–523.
-
- White TR, Renzelman CM, Rand AC, Rezai T, McEwen CM, Gelev VM, Turner RA, Linington RG, Leung SSF, Kalgutkar AS, Bauman JN, Zhang YZ, Liras S, Price DA, Mathiowetz AM, Jacobson MP, Lokey RS. Nat Chem Biol. 2011;7:810–817. - PMC - PubMed
- Rezai T, Yu B, Millhauser GL, Jacobson MP, Lokey RS. J Am Chem Soc. 2006;128:2510–2511. - PubMed
- Fairlie DP, Abbenante G, March DR. Curr Med Chem. 1995;2:654–686.
- Kwon YU, Kodadek T. Chem Biol. 2007;14:671–677. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

